+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13093 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
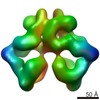
| Title | Immature 60S Ribosomal Subunit from C. thermophilum | |||||||||
 Map data Map data | Immature 60S Ribosomal Subunit from C.thermophilum, Main Map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) | |||||||||
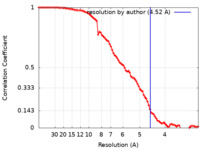
| Method | single particle reconstruction / cryo EM / Resolution: 4.52 Å | |||||||||
 Authors Authors | Skalidis I / Kastritis PL | |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Cryo-EM and artificial intelligence visualize endogenous protein community members. Authors: Ioannis Skalidis / Fotis L Kyrilis / Christian Tüting / Farzad Hamdi / Grzegorz Chojnowski / Panagiotis L Kastritis /  Abstract: Cellular function is underlined by megadalton assemblies organizing in proximity, forming communities. Metabolons are protein communities involving metabolic pathways such as protein, fatty acid, and ...Cellular function is underlined by megadalton assemblies organizing in proximity, forming communities. Metabolons are protein communities involving metabolic pathways such as protein, fatty acid, and thioesters of coenzyme-A synthesis. Metabolons are highly heterogeneous due to their function, making their analysis particularly challenging. Here, we simultaneously characterize metabolon-embedded architectures of a 60S pre-ribosome, fatty acid synthase, and pyruvate/oxoglutarate dehydrogenase complex E2 cores de novo. Cryo-electron microscopy (cryo-EM) 3D reconstructions are resolved at 3.84-4.52 Å resolution by collecting <3,000 micrographs of a single cellular fraction. After combining cryo-EM with artificial intelligence-based atomic modeling and de novo sequence identification methods, at this resolution range, polypeptide hydrogen bonding patterns are discernible. Residing molecular components resemble their purified counterparts from other eukaryotes but also exhibit substantial conformational variation with potential functional implications. Our results propose an integrated tool, boosted by machine learning, that opens doors for structural systems biology spearheaded by cryo-EM characterization of native cell extracts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13093.map.gz emd_13093.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13093-v30.xml emd-13093-v30.xml emd-13093.xml emd-13093.xml | 31.4 KB 31.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13093_fsc.xml emd_13093_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_13093.png emd_13093.png | 61.1 KB | ||
| Others |  emd_13093_additional_1.map.gz emd_13093_additional_1.map.gz emd_13093_half_map_1.map.gz emd_13093_half_map_1.map.gz emd_13093_half_map_2.map.gz emd_13093_half_map_2.map.gz | 117.2 MB 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13093 http://ftp.pdbj.org/pub/emdb/structures/EMD-13093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13093 | HTTPS FTP |
-Related structure data
| Related structure data |  7q5qC  7q5rC  7q5sC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10892 (Title: Cryo-EM SPA dataset of Megadalton-range protein communities from a Chaetomium thermophilum native cell extract EMPIAR-10892 (Title: Cryo-EM SPA dataset of Megadalton-range protein communities from a Chaetomium thermophilum native cell extractData size: 1.1 TB Data #1: Unaligned fractions saved by Falcon 3 EC camera [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13093.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13093.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Immature 60S Ribosomal Subunit from C.thermophilum, Main Map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5678 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Signature 4 Map
| File | emd_13093_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Signature 4 Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Immature 60S Ribosomal Subunit from C.thermophilum, Half-Map A
| File | emd_13093_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Immature 60S Ribosomal Subunit from C.thermophilum, Half-Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Immature 60S Ribosomal Subunit from C.thermophilum, Half-Map B
| File | emd_13093_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Immature 60S Ribosomal Subunit from C.thermophilum, Half-Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Native 60-mer core of Pyruvate Dehydrogenase Complex
+Supramolecule #1: Native 60-mer core of Pyruvate Dehydrogenase Complex
+Macromolecule #1: uL13
+Macromolecule #2: eL21
+Macromolecule #3: eL13
+Macromolecule #4: eL15
+Macromolecule #5: eL18
+Macromolecule #6: uL15
+Macromolecule #7: eL36
+Macromolecule #8: eL32
+Macromolecule #9: eL20
+Macromolecule #10: uL22
+Macromolecule #11: eL8
+Macromolecule #12: uL29
+Macromolecule #13: uL6
+Macromolecule #14: eL6
+Macromolecule #15: uL5
+Macromolecule #16: uL18
+Macromolecule #17: eL14
+Macromolecule #18: uL30
+Macromolecule #19: eL33
+Macromolecule #20: uL24
+Macromolecule #21: uL4
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 200.0 mM / Component - Formula: NH4CH2COOH / Component - Name: Ammonium acetate |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: For plunging, blot force 0 and blotting time of 4 sec were applied.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Temperature | Min: 77.15 K / Max: 103.15 K |
| Alignment procedure | Coma free - Residual tilt: 14.7 mrad |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 2808 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 92000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)