+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13043 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | La Crosse virus polymerase at replication initiation stage | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA-dependent RNA polymerase / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum / virion component / host cell endoplasmic reticulum-Golgi intermediate compartment / host cell Golgi apparatus / Hydrolases; Acting on ester bonds / hydrolase activity / RNA-directed RNA polymerase / viral RNA genome replication / nucleotide binding / RNA-directed RNA polymerase activity ...host cell endoplasmic reticulum / virion component / host cell endoplasmic reticulum-Golgi intermediate compartment / host cell Golgi apparatus / Hydrolases; Acting on ester bonds / hydrolase activity / RNA-directed RNA polymerase / viral RNA genome replication / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  La Crosse orthobunyavirus La Crosse orthobunyavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Arragain B / Durieux Trouilleton Q | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural snapshots of La Crosse virus polymerase reveal the mechanisms underlying Peribunyaviridae replication and transcription. Authors: Benoît Arragain / Quentin Durieux Trouilleton / Florence Baudin / Jan Provaznik / Nayara Azevedo / Stephen Cusack / Guy Schoehn / Hélène Malet /   Abstract: Segmented negative-strand RNA bunyaviruses encode a multi-functional polymerase that performs genome replication and transcription. Here, we establish conditions for in vitro activity of La Crosse ...Segmented negative-strand RNA bunyaviruses encode a multi-functional polymerase that performs genome replication and transcription. Here, we establish conditions for in vitro activity of La Crosse virus polymerase and visualize its conformational dynamics by cryo-electron microscopy, unveiling the precise molecular mechanics underlying its essential activities. We find that replication initiation is coupled to distal duplex promoter formation, endonuclease movement, prime-and-realign loop extension and closure of the polymerase core that direct the template towards the active site. Transcription initiation depends on C-terminal region closure and endonuclease movements that prompt primer cleavage prior to primer entry in the active site. Product realignment after priming, observed in replication and transcription, is triggered by the prime-and-realign loop. Switch to elongation results in polymerase reorganization and core region opening to facilitate template-product duplex formation in the active site cavity. The uncovered detailed mechanics should be helpful for the future design of antivirals counteracting bunyaviral life threatening pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13043.map.gz emd_13043.map.gz | 25.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13043-v30.xml emd-13043-v30.xml emd-13043.xml emd-13043.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
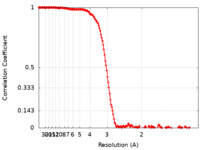
| FSC (resolution estimation) |  emd_13043_fsc.xml emd_13043_fsc.xml | 15.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13043.png emd_13043.png | 110.3 KB | ||
| Filedesc metadata |  emd-13043.cif.gz emd-13043.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13043 http://ftp.pdbj.org/pub/emdb/structures/EMD-13043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13043 | HTTPS FTP |
-Related structure data
| Related structure data |  7ornMC  7oriC  7orjC  7orkC  7orlC  7ormC  7oroC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11021 (Title: Cryo-EM data used for the determination of the structures of LACV-L in 3 different states: replication initiation state, transcription capped primer active site entry state and transcription initiation state EMPIAR-11021 (Title: Cryo-EM data used for the determination of the structures of LACV-L in 3 different states: replication initiation state, transcription capped primer active site entry state and transcription initiation stateData size: 2.2 TB Data #1: Unaligned multiframe micrographs of LACV-L incubated with capped RNA 14-mer, 3' vRNA 1-25, 5' 1-17BPm, UTP, ATP, MgCl2 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13043.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13043.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.645 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : La Crosse virus polymerase at replication initiation
| Entire | Name: La Crosse virus polymerase at replication initiation |
|---|---|
| Components |
|
-Supramolecule #1: La Crosse virus polymerase at replication initiation
| Supramolecule | Name: La Crosse virus polymerase at replication initiation / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 279 KDa |
-Supramolecule #2: La Crosse virus polymerase
| Supramolecule | Name: La Crosse virus polymerase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  La Crosse orthobunyavirus La Crosse orthobunyavirus |
-Supramolecule #3: RNA
| Supramolecule | Name: RNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  La Crosse orthobunyavirus La Crosse orthobunyavirus |
-Macromolecule #1: La Crosse virus polymerase
| Macromolecule | Name: La Crosse virus polymerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  La Crosse orthobunyavirus La Crosse orthobunyavirus |
| Molecular weight | Theoretical: 264.751062 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDYQEYQQFL ARINTARDAC VAKDIDVDLL MARKDYFGRE LCKSLNIEYR NDVPFIDIIL DIRPEVDPLT IDAPHITPDN YLYINNVLY IIDYKVSVSN ESSVITYDKY YELTRDISDR LSIPIEIVII RIDPVSRDLH INSDRFKELY PTIVVDINFN Q FFDLKQLL ...String: MDYQEYQQFL ARINTARDAC VAKDIDVDLL MARKDYFGRE LCKSLNIEYR NDVPFIDIIL DIRPEVDPLT IDAPHITPDN YLYINNVLY IIDYKVSVSN ESSVITYDKY YELTRDISDR LSIPIEIVII RIDPVSRDLH INSDRFKELY PTIVVDINFN Q FFDLKQLL YEKFGDDEEF LLKVAHGDFT LTAPWCKTGC PEFWKHPIYK EFKMSMPVPE RRLFEESVKF NAYESERWNT NL VKIREYT KKDYSEHISK SAKNIFLASG FYKQPNKNEI SEGWTLMVER VQDQREISKS LHDQKPSIHF IWGAHNPGNS NNA TFKLIL LSKSLQSIKG ISTYTEAFKS LGKMMDIGDK AIEYEEFCMS LKSKARSSWK QIMNKKLEPK QINNALVLWE QQFM INNDL IDKSEKLKLF KNFCGIGKHK QFKNKMLEDL EVSKPKILDF DDANMYLASL TMMEQSKKIL SKSNGLKPDN FILNE FGSR IKDANKETYD NMHKIFETGY WQCISDFSTL MKNILSVSQY NRHNTFRIAM CANNNVFAIV FPSADIKTKK ATVVYS IIV LHKEEENIFN PGCLHGTFKC MNGYISISRA IRLDKERCQR IVSSPGLFLT TCLLFKHDNP TLVMSDIMNF SIYTSLS IT KSVLSLTEPA RYMIMNSLAI SSNVKDYIAE KFSPYTKTLF SVYMTRLIKN ACFDAYDQRQ RVQLRDIYLS DYDITQKG I KDNRELTSIW FPGSVTLKEY LTQIYLPFYF NAKGLHEKHH VMVDLAKTIL EIECEQRENI KEIWSTNCTK QTVNLKILI HSLCKNLLAD TSRHNHLRNR IENRNNFRRS ITTISTFTSS KSCLKIGDFR KEKELQSVKQ KKILEVQSRK MRLANPMFVT DEQVCLEVG HCNYEMLRNA MPNYTDYIST KVFDRLYELL DKKVLTDKPV IEQIMDMMID HKKFYFTFFN KGQKTSKDRE I FVGEYEAK MCMYAVERIA KERCKLNPDE MISEPGDGKL KVLEQKSEQE IRFLVETTRQ KNREIDEAIE ALATEGSGWS HP QFEKGSG YESNLGKIEK LSLGKAKGLK MEINADMSKW SAQDVFYKYF WLIALDPILY PQEKERILYF MCNYMDKELI LPD ELLFNL LDQKVAYQND IIATMTNQLN SNTVLIKRNW LQGNFNYTSS YVHSCAMSVY KEILKEAITL LDGSILVNSL VHSD DNQTS ITIVQDKMEN DKIIDFAMKE FERACLTFGC QANMKKTYVT NCIKEFVSLF NLYGEPFSIY GRFLLTSVGD CAYIG PYED LASRISSAQT AIKHGCPPSL AWVSIAISHW MTSLTYNMLP GQSNDPIDYF PAENRKDIPI ELNGVLDAPL SMISTV GLE SGNLYFLIKL LSKYTPVMQK RESVVNQIAE VKNWKVEDLT DNEIFRLKIL RYLVLDAEMD PSDIMGETSD MRGRSIL TP RKFTTAGSLR KLYSFSKYQD RLSSPGGMVE LFTYLLEKPE LLVTKGEDMK DYMESVIFRY NSKRFKESLS IQNPAQLF I EQILFSHKPV IDFSGIRDKY INLHDSRALE KEPDILGKVT FTEAYRLLMR DLSSLELTND DIQVIYSYII LNDPMMITI ANTHILSIYG SPQRRMGMSC STMPEFRNLK LIHHSPALVL RAYSKNNPDI QGADPTEMAR DLVHLKEFVE NTNLEEKMKV RIAMNEAEK GQRDIVFELK EMTRFYQVCY EYVKSTEHKI KVFILPAKSY TTTDFCSLMQ GNLIKDKEWY TVHYLKQILS G GHKAIMQH NATSEQNIAF ECFKLITHFA DSFIDSLSRS AFLQLIIDEF SYKDVKVSKL YDIIKNGYNR TDFIPLLFRT GD LRQADLD KYDAMKSHER VTWNDWQTSR HLDMGSINLT ITGYNRSITI IGEDNKLTYA ELCLTRKTPE NITISGRKLL GSR HGLKFE NMSKIQTYPG NYYITYRKKD RHQFVYQIHS HESITRRNEE HMAIRTRIYN EITPVCVVNV AEVDGDQRIL IRSL DYLNN DIFSLSRIKV GLDEFATIKK AHFSKMVSFE GPPIKTGLLD LTELMKSQDL LNLNYDNIRN SNLISFSKLI CCEGS DNIN DGLEFLSDDP MNFTEGEAIH STPIFNIYYS KRGERHMTYR NAIKLLIERE TKIFEEAFTF SENGFISPEN LGCLEA VVS LIKLLKTNEW STVIDKCIHI CLIKNGMDHM YHSFDVPKCF MGNPITRDIN WVMFREFINS LPGTDIPPWN VMTENFK KK CIALINSKFE TQRDFSEFTK LMKKEGGRSN IEFD UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*CP*AP*AP*G)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*CP*GP*AP*GP*UP*GP*UP*CP*GP*UP*AP*CP*CP*AP*AP*G)-3') type: rna / ID: 2 Details: Mutated 5' sequence of La Crosse virus M segment Nucleotides G2, U3, A9 and C10 were mutated into C2, G3, C9 and G10 Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  La Crosse orthobunyavirus La Crosse orthobunyavirus |
| Molecular weight | Theoretical: 5.466325 KDa |
| Sequence | String: ACGAGUGUCG UACCAAG |
-Macromolecule #3: RNA (5'-R(P*AP*CP*UP*UP*GP*GP*UP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*AP*CP*UP*UP*GP*GP*UP*AP*GP*UP*AP*CP*AP*CP*UP*AP*CP*U)-3') type: rna / ID: 3 Details: 3' extremity (25 nucleotides) of La Crosse virus M segment Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  La Crosse orthobunyavirus La Crosse orthobunyavirus |
| Molecular weight | Theoretical: 7.882668 KDa |
| Sequence | String: UAUCUAUACU UGGUAGUACA CUACU |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 92 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 50 mM TRIS-HCl pH 8, 150 mM NaCl, 5 mM BME, 100 uM ATP/UTP and 5mM MgCl2. | |||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Details: 25 mA | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: blot force 1 2 sec. | |||||||||||||||||||||
| Details | 1.3 uM LACV-LCItag_H34K were sequentially incubated for 1h at 4 degree with (i) 1.9 uM 5prime 1-17 BPm, (ii) 1.9 uM 3prime vRNA 1-25. 100 uM ATP/UTP and 5mM MgCl2 for 4h at 30dregree |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 15573 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 18.0 µm / Calibrated defocus min: 0.8 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 18.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)