+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12531 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T20S proteasome subtomogram average | |||||||||
 Map data Map data | T20S proteasome subtomogram average | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Fernandez JJ | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2018 Journal: J Struct Biol / Year: 2018Title: Cryo-tomography tilt-series alignment with consideration of the beam-induced sample motion. Authors: Jose-Jesus Fernandez / Sam Li / Tanmay A M Bharat / David A Agard /    Abstract: Recent evidence suggests that the beam-induced motion of the sample during tilt-series acquisition is a major resolution-limiting factor in electron cryo-tomography (cryoET). It causes suboptimal ...Recent evidence suggests that the beam-induced motion of the sample during tilt-series acquisition is a major resolution-limiting factor in electron cryo-tomography (cryoET). It causes suboptimal tilt-series alignment and thus deterioration of the reconstruction quality. Here we present a novel approach to tilt-series alignment and tomographic reconstruction that considers the beam-induced sample motion through the tilt-series. It extends the standard fiducial-based alignment approach in cryoET by introducing quadratic polynomials to model the sample motion. The model can be used during reconstruction to yield a motion-compensated tomogram. We evaluated our method on various datasets with different sample sizes. The results demonstrate that our method could be a useful tool to improve the quality of tomograms and the resolution in cryoET. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12531.map.gz emd_12531.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12531-v30.xml emd-12531-v30.xml emd-12531.xml emd-12531.xml | 7.2 KB 7.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12531.png emd_12531.png | 128.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12531 http://ftp.pdbj.org/pub/emdb/structures/EMD-12531 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12531 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12531 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10651 (Title: CryoET dataset of T20S proteasome for testing motion-aware tilt-series alignment and 3D reconstruction EMPIAR-10651 (Title: CryoET dataset of T20S proteasome for testing motion-aware tilt-series alignment and 3D reconstructionData size: 5.1 Data #1: Aligned tilt-series of T20S proteasome [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12531.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12531.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T20S proteasome subtomogram average | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.56 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
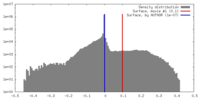
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : T20S proteasome
| Entire | Name: T20S proteasome |
|---|---|
| Components |
|
-Supramolecule #1: T20S proteasome
| Supramolecule | Name: T20S proteasome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: D7 (2x7 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 9.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number subtomograms used: 3928 |
|---|---|
| Extraction | Number tomograms: 14 / Number images used: 3928 |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)