+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10737 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
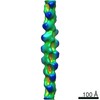
| Title | Actin filament structure from vinculin-induced bundles | |||||||||
 Map data Map data | Actin filament structure from vinculin-induced bundles | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 14.2 Å | |||||||||
 Authors Authors | Boujemaa-Paterski R / Martins B / Eibauer M / Geiger B / Medalia O | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Talin-activated vinculin interacts with branched actin networks to initiate bundles. Authors: Rajaa Boujemaa-Paterski / Bruno Martins / Matthias Eibauer / Charlie T Beales / Benjamin Geiger / Ohad Medalia /    Abstract: Vinculin plays a fundamental role in integrin-mediated cell adhesion. Activated by talin, it interacts with diverse adhesome components, enabling mechanical coupling between the actin cytoskeleton ...Vinculin plays a fundamental role in integrin-mediated cell adhesion. Activated by talin, it interacts with diverse adhesome components, enabling mechanical coupling between the actin cytoskeleton and the extracellular matrix. Here we studied the interactions of activated full-length vinculin with actin and the way it regulates the organization and dynamics of the Arp2/3 complex-mediated branched actin network. Through a combination of surface patterning and light microscopy experiments we show that vinculin can bundle dendritic actin networks through rapid binding and filament crosslinking. We show that vinculin promotes stable but flexible actin bundles having a mixed-polarity organization, as confirmed by cryo-electron tomography. Adhesion-like synthetic design of vinculin activation by surface-bound talin revealed that clustered vinculin can initiate and immobilize bundles from mobile Arp2/3-branched networks. Our results provide a molecular basis for coordinate actin bundle formation at nascent adhesions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10737.map.gz emd_10737.map.gz | 8.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10737-v30.xml emd-10737-v30.xml emd-10737.xml emd-10737.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10737.png emd_10737.png | 29 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10737 http://ftp.pdbj.org/pub/emdb/structures/EMD-10737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10737 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10737 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10548 (Title: Cryo-ET of actin bundles induced by full length vinculin EMPIAR-10548 (Title: Cryo-ET of actin bundles induced by full length vinculinData size: 4.0 Data #1: Tomograms of actin bundles induced by full length vinculin [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10737.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10737.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actin filament structure from vinculin-induced bundles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.443 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Actin
| Entire | Name: Actin |
|---|---|
| Components |
|
-Supramolecule #1: Actin
| Supramolecule | Name: Actin / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin
| Macromolecule | Name: Actin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTL KYPIEHGIIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE K MTQIMFET FNVPAMYVAI QAVLSLYASG RTTGIVLDSG DGVTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTL KYPIEHGIIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE K MTQIMFET FNVPAMYVAI QAVLSLYASG RTTGIVLDSG DGVTHNVPIY EGYALPHAIM RL DLAGRDL TDYLMKILTE RGYSFVTTAE REIVRDIKEK LCYVALDFEN EMATAASSSS LEK SYELPD GQVITIGNER FRCPETLFQP SFIGMESAGI HETTYNSIMK CDIDIRKDLY ANNV MSGGT TMYPGIADRM QKEITALAPS TMKIKIIAPP ERKYSVWIGG SILASLSTFQ QMWIT KQEY DEAGPSIVHR KCF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 1.2 sec. / Average electron dose: 1.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 42000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 3.5 µm / Nominal magnification: 14522 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















