+ Open data
Open data
- Basic information
Basic information
| Entry | 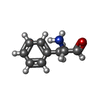 Database: PDB chemical components / ID: PHA Database: PDB chemical components / ID: PHA |
|---|---|
| Name | Name: |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: L-PEPTIDE LINKING / PDB classification: ATOMP / One letter code: F / Three letter code: PHA / Model coordinates PDB-ID: 3SGA / Parent comp.: PHE | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Nikkaji / Nikkaji /  PubChem / PubChem /  SureChEMBL / SureChEMBL /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | (| OpenEye OEToolkits 1.5.0 | ( | |
|---|
-PDB entries
Showing all 4 items

PDB-1kqs: 
The Haloarcula marismortui 50S Complexed with a Pretranslocational Intermediate in Protein Synthesis

PDB-1m90: 
Co-crystal structure of CCA-Phe-caproic acid-biotin and sparsomycin bound to the 50S ribosomal subunit

PDB-1q86: 
Crystal structure of CCA-Phe-cap-biotin bound simultaneously at half occupancy to both the A-site and P-site of the the 50S ribosomal Subunit.

PDB-4v68: 
T. thermophilus 70S ribosome in complex with mRNA, tRNAs and EF-Tu.GDP.kirromycin ternary complex, fitted to a 6.4 A Cryo-EM map.
 Movie
Movie Controller
Controller