[English] 日本語
 Yorodumi
Yorodumi- ChemComp-031: (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benz... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 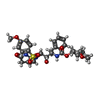 Database: PDB chemical components / ID: 031 Database: PDB chemical components / ID: 031 |
|---|---|
| Name | Name: ( |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: 31 / Ideal coordinates details: Corina / Model coordinates PDB-ID: 3H5B | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  BindingDB / BindingDB /  Brenda / Brenda /  ChEMBL / ChEMBL /  Nikkaji / Nikkaji /  PubChem / PubChem /  PubChem_TPharma PubChem_TPharma  / /  SureChEMBL / SureChEMBL /  ZINC / ZINC /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | (| OpenEye OEToolkits 1.5.0 | [( | |
|---|
-PDB entries
Showing all 6 items

PDB-3h5b: 
Crystal structure of wild type HIV-1 protease with novel P1'-ligand GRL-02031

PDB-3vf5: 
Crystal Structure of HIV-1 Protease Mutant I47V with novel P1'-Ligands GRL-02031
PDB-3vf7: 
Crystal Structure of HIV-1 Protease Mutant L76V with novel P1'-Ligands GRL-02031

PDB-3vfa: 
Crystal Structure of HIV-1 Protease Mutant V82A with novel P1'-Ligands GRL-02031

PDB-3vfb: 
Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031

PDB-4j55: 
Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical isolate PR20 with the potent antiviral inhibitor GRL-02031
 Movie
Movie Controller
Controller


