+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9392 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage L virion | |||||||||
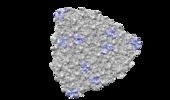
 Map data Map data | 1/8th of the virion for phage L-segmentation shows capsid protein only | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Dec protein Function and homology information Function and homology information | |||||||||
| Biological species |  Enterobacteria phage L (virus) Enterobacteria phage L (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Parent KN / Schrad JR | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: The phage L capsid decoration protein has a novel OB-fold and an unusual capsid binding strategy. Authors: Rebecca L Newcomer / Jason R Schrad / Eddie B Gilcrease / Sherwood R Casjens / Michael Feig / Carolyn M Teschke / Andrei T Alexandrescu / Kristin N Parent /  Abstract: The major coat proteins of dsDNA tailed phages (order ) and herpesviruses form capsids by a mechanism that includes active packaging of the dsDNA genome into a precursor procapsid, followed by ...The major coat proteins of dsDNA tailed phages (order ) and herpesviruses form capsids by a mechanism that includes active packaging of the dsDNA genome into a precursor procapsid, followed by expansion and stabilization of the capsid. These viruses have evolved diverse strategies to fortify their capsids, such as non-covalent binding of auxiliary 'decoration' (Dec) proteins. The Dec protein from the P22-like phage L has a highly unusual binding strategy that distinguishes between nearly identical three-fold and quasi-three-fold sites of the icosahedral capsid. Cryo-electron microscopy and three-dimensional image reconstruction were employed to determine the structure of native phage L particles. NMR was used to determine the structure/dynamics of Dec in solution. The NMR structure and the cryo-EM density envelope were combined to build a model of the capsid-bound Dec trimer. Key regions that modulate the binding interface were verified by site-directed mutagenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9392.map.gz emd_9392.map.gz | 64.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9392-v30.xml emd-9392-v30.xml emd-9392.xml emd-9392.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9392.png emd_9392.png | 272.3 KB | ||
| Others |  emd_9392_additional.map.gz emd_9392_additional.map.gz | 688.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9392 http://ftp.pdbj.org/pub/emdb/structures/EMD-9392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9392 | HTTPS FTP |
-Validation report
| Summary document |  emd_9392_validation.pdf.gz emd_9392_validation.pdf.gz | 77.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9392_full_validation.pdf.gz emd_9392_full_validation.pdf.gz | 76.9 KB | Display | |
| Data in XML |  emd_9392_validation.xml.gz emd_9392_validation.xml.gz | 492 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9392 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9392 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9392.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9392.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 1/8th of the virion for phage L-segmentation shows capsid protein only | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
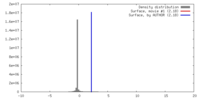
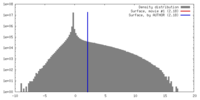
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: single Dec trimer segmented from phage L virion map
| File | emd_9392_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | single Dec trimer segmented from phage L virion map | ||||||||||||
| Projections & Slices |
| ||||||||||||
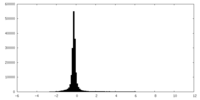
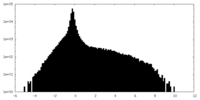
| Density Histograms |
- Sample components
Sample components
-Entire : Enterobacteria phage L
| Entire | Name:  Enterobacteria phage L (virus) Enterobacteria phage L (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage L
| Supramolecule | Name: Enterobacteria phage L / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 45441 / Sci species name: Enterobacteria phage L / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
| Molecular weight | Theoretical: 60 MDa |
| Virus shell | Shell ID: 1 / Name: coat protein / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 100.0 nm / Details: unspecified | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER | |||||||||
| Details | concentration is 1x10^12 phage/mL |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 5000 pixel / Digitization - Dimensions - Height: 4000 pixel / Digitization - Sampling interval: 6.4 µm / Digitization - Frames/image: 0-53 / Number grids imaged: 1 / Number real images: 494 / Average exposure time: 2.0 sec. / Average electron dose: 27.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.49 µm / Nominal defocus min: 0.35 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)