+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9028 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mtb ClpB in complex with AMPPNP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryoEM / pathogen / AAA ATPase / Mycobacterium tuberculosis / unfoldase / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidoglycan-based cell wall / cellular response to heat / protein refolding / ATP hydrolysis activity / ATP binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
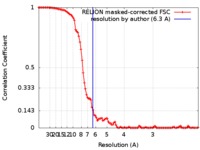
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Yu HJ / Li HL | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2018 Journal: Proc Natl Acad Sci U S A / Year: 2018Title: ATP hydrolysis-coupled peptide translocation mechanism of ClpB. Authors: Hongjun Yu / Tania J Lupoli / Amanda Kovach / Xing Meng / Gongpu Zhao / Carl F Nathan / Huilin Li /  Abstract: The protein disaggregase ClpB hexamer is conserved across evolution and has two AAA+-type nucleotide-binding domains, NBD1 and NBD2, in each protomer. In (), ClpB facilitates asymmetric distribution ...The protein disaggregase ClpB hexamer is conserved across evolution and has two AAA+-type nucleotide-binding domains, NBD1 and NBD2, in each protomer. In (), ClpB facilitates asymmetric distribution of protein aggregates during cell division to help the pathogen survive and persist within the host, but a mechanistic understanding has been lacking. Here we report cryo-EM structures at 3.8- to 3.9-Å resolution of ClpB bound to a model substrate, casein, in the presence of the weakly hydrolyzable ATP mimic adenosine 5'-[γ-thio]triphosphate. ClpB existed in solution in two closed-ring conformations, conformers 1 and 2. In both conformers, the 12 pore-loops on the 12 NTDs of the six protomers (P1-P6) were arranged similarly to a staircase around the bound peptide. Conformer 1 is a low-affinity state in which three of the 12 pore-loops (the protomer P1 NBD1 and NBD2 loops and the protomer P2 NBD1 loop) are not engaged with peptide. Conformer 2 is a high-affinity state because only one pore-loop (the protomer P2 NBD1 loop) is not engaged with the peptide. The resolution of the two conformations, along with their bound substrate peptides and nucleotides, enabled us to propose a nucleotide-driven peptide translocation mechanism of a bacterial ClpB that is largely consistent with several recent unfoldase structures, in particular with the eukaryotic Hsp104. However, whereas Hsp104's two NBDs move in opposing directions during one step of peptide translocation, in Mtb ClpB the two NBDs move only in the direction of translocation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9028.map.gz emd_9028.map.gz | 88.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9028-v30.xml emd-9028-v30.xml emd-9028.xml emd-9028.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9028_fsc.xml emd_9028_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_9028.png emd_9028.png | 116.3 KB | ||
| Filedesc metadata |  emd-9028.cif.gz emd-9028.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9028 http://ftp.pdbj.org/pub/emdb/structures/EMD-9028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9028 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9028 | HTTPS FTP |
-Related structure data
| Related structure data |  6ed3MC  7942C  7943C  9027C  6djuC  6djvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9028.map.gz / Format: CCP4 / Size: 95 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9028.map.gz / Format: CCP4 / Size: 95 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.074 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mtb ClpB in complex with AMPPNP
| Entire | Name: Mtb ClpB in complex with AMPPNP |
|---|---|
| Components |
|
-Supramolecule #1: Mtb ClpB in complex with AMPPNP
| Supramolecule | Name: Mtb ClpB in complex with AMPPNP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chaperone protein ClpB
| Macromolecule | Name: Chaperone protein ClpB / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.688281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSFNPTTKT QAALTAALQA ASTAGNPEIR PAHLLMALLT QNDGIAAPLL EAVGVEPATV RAETQRLLDR LPQATGASTQ PQLSRESLA AITTAQQLAT ELDDEYVSTE HVMVGLATGD SDVAKLLTGH GASPQALREA FVKVRGSARV TSPEPEATYQ A LQKYSTDL ...String: MDSFNPTTKT QAALTAALQA ASTAGNPEIR PAHLLMALLT QNDGIAAPLL EAVGVEPATV RAETQRLLDR LPQATGASTQ PQLSRESLA AITTAQQLAT ELDDEYVSTE HVMVGLATGD SDVAKLLTGH GASPQALREA FVKVRGSARV TSPEPEATYQ A LQKYSTDL TARAREGKLD PVIGRDNEIR RVVQVLSRRT KNNPVLIGEP GVGKTAIVEG LAQRIVAGDV PESLRDKTIV AL DLGSMVA GSKYRGEFEE RLKAVLDDIK NSAGQIITFI DELHTIVGAG ATGEGAMDAG NMIKPMLARG ELRLVGATTL DEY RKHIEK DAALERRFQQ VYVGEPSVED TIGILRGLKD RYEVHHGVRI TDSALVAAAT LSDRYITARF LPDKAIDLVD EAAS RLRME IDSRPVEIDE VERLVRRLEI EEMALSKEED EASAERLAKL RSELADQKEK LAELTTRWQN EKNAIEIVRD LKEQL EALR GESERAERDG DLAKAAELRY GRIPEVEKKL DAALPQAQAR EQVMLKEEVG PDDIADVVSA WTGIPAGRLL EGETAK LLR MEDELGKRVI GQKAAVTAVS DAVRRSRAGV SDPNRPTGAF MFLGPTGVGK TELAKALADF LFDDERAMVR IDMSEYG EK HTVARLIGAP PGYVGYEAGG QLTEAVRRRP YTVVLFDEIE KAHPDVFDVL LQVLDEGRLT DGHGRTVDFR NTILILTS N LGSGGSAEQV LAAVRATFKP EFINRLDDVL IFEGLNPEEL VRIVDIQLAQ LGKRLAQRRL QLQVSLPAKR WLAQRGFDP VYGARPLRRL VQQAIGDQLA KMLLAGQVHD GDTVPVNVSP DADSLILG UniProtKB: Chaperone protein ClpB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)