+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8077 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MicroED structure of proteinase K at 1.75 A resolution | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidase K / serine-type endopeptidase activity / proteolysis / extracellular region / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Engyodontium album (fungus) Engyodontium album (fungus) | |||||||||
| Method | electron crystallography / cryo EM / Resolution: 1.75 Å | |||||||||
 Authors Authors | Hattne J / Shi D | |||||||||
 Citation Citation |  Journal: J Appl Crystallogr / Year: 2016 Journal: J Appl Crystallogr / Year: 2016Title: Modeling truncated pixel values of faint reflections in MicroED images. Authors: Johan Hattne / Dan Shi / M Jason de la Cruz / Francis E Reyes / Tamir Gonen /  Abstract: The weak pixel counts surrounding the Bragg spots in a diffraction image are important for establishing a model of the background underneath the peak and estimating the reliability of the integrated ...The weak pixel counts surrounding the Bragg spots in a diffraction image are important for establishing a model of the background underneath the peak and estimating the reliability of the integrated intensities. Under certain circumstances, particularly with equipment not optimized for low-intensity measurements, these pixel values may be corrupted by corrections applied to the raw image. This can lead to truncation of low pixel counts, resulting in anomalies in the integrated Bragg intensities, such as systematically higher signal-to-noise ratios. A correction for this effect can be approximated by a three-parameter lognormal distribution fitted to the weakly positive-valued pixels at similar scattering angles. The procedure is validated by the improved refinement of an atomic model against structure factor amplitudes derived from corrected micro-electron diffraction (MicroED) images. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8077.map.gz emd_8077.map.gz | 3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8077-v30.xml emd-8077-v30.xml emd-8077.xml emd-8077.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8077.png emd_8077.png | 206.5 KB | ||
| Filedesc metadata |  emd-8077.cif.gz emd-8077.cif.gz | 5.5 KB | ||
| Filedesc structureFactors |  emd_8077_sf.cif.gz emd_8077_sf.cif.gz | 2.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8077 http://ftp.pdbj.org/pub/emdb/structures/EMD-8077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8077 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8077 | HTTPS FTP |
-Validation report
| Summary document |  emd_8077_validation.pdf.gz emd_8077_validation.pdf.gz | 635.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8077_full_validation.pdf.gz emd_8077_full_validation.pdf.gz | 635.5 KB | Display | |
| Data in XML |  emd_8077_validation.xml.gz emd_8077_validation.xml.gz | 4.3 KB | Display | |
| Data in CIF |  emd_8077_validation.cif.gz emd_8077_validation.cif.gz | 4.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8077 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8077 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8077 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8077 | HTTPS FTP |
-Related structure data
| Related structure data |  5i9sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8077.map.gz / Format: CCP4 / Size: 3.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8077.map.gz / Format: CCP4 / Size: 3.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.60105 Å / Y: 0.60105 Å / Z: 0.5739 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
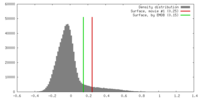
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 96 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Proteinase K
| Entire | Name: Proteinase K |
|---|---|
| Components |
|
-Supramolecule #1: Proteinase K
| Supramolecule | Name: Proteinase K / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Engyodontium album (fungus) Engyodontium album (fungus) |
| Molecular weight | Theoretical: 28.98 KDa |
-Macromolecule #1: Proteinase K
| Macromolecule | Name: Proteinase K / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: peptidase K |
|---|---|
| Source (natural) | Organism:  Engyodontium album (fungus) Engyodontium album (fungus) |
| Molecular weight | Theoretical: 28.958791 KDa |
| Sequence | String: AAQTNAPWGL ARISSTSPGT STYYYDESAG QGSCVYVIDT GIEASHPEFE GRAQMVKTYY YSSRDGNGHG THCAGTVGSR TYGVAKKTQ LFGVKVLDDN GSGQYSTIIA GMDFVASDKN NRNCPKGVVA SLSLGGGYSS SVNSAAARLQ SSGVMVAVAA G NNNADARN ...String: AAQTNAPWGL ARISSTSPGT STYYYDESAG QGSCVYVIDT GIEASHPEFE GRAQMVKTYY YSSRDGNGHG THCAGTVGSR TYGVAKKTQ LFGVKVLDDN GSGQYSTIIA GMDFVASDKN NRNCPKGVVA SLSLGGGYSS SVNSAAARLQ SSGVMVAVAA G NNNADARN YSPASEPSVC TVGASDRYDR RSSFSNYGSV LDIFGPGTDI LSTWIGGSTR SISGTSMATP HVAGLAAYLM TL GKTTAAS ACRYIADTAN KGDLSNIPFG TVNLLAYNNY QA UniProtKB: Proteinase K |
-Macromolecule #2: SULFATE ION
| Macromolecule | Name: SULFATE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: SO4 |
|---|---|
| Molecular weight | Theoretical: 96.063 Da |
| Chemical component information |  ChemComp-SO4: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 133 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 25 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 / Component:
| ||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number grids imaged: 1 / Number real images: 184 / Number diffraction images: 184 / Average exposure time: 4.1 sec. / Average electron dose: 0.004 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Camera length: 1200 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 1-279 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Electron scattering factors |
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 17.2 |
| Output model |  PDB-5i9s: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)