+ Open data
Open data
- Basic information
Basic information
| Entry | 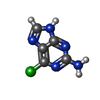 Database: PDB chemical components / ID: 6GU Database: PDB chemical components / ID: 6GU |
|---|---|
| Name | Name: |
-Chemical information
| Composition |  | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: 6GU / Ideal coordinates details: Corina / Model coordinates PDB-ID: 3E9Z | ||||||||
| History |
| ||||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  BindingDB BindingDB       / /  Brenda Brenda   / /  ChEBI / ChEBI /  ChEMBL / ChEMBL /  ChemicalBook / ChemicalBook /  CompTox / CompTox /  HMDB / HMDB /  Nikkaji / Nikkaji /  PubChem / PubChem /  PubChem_TPharma PubChem_TPharma  / /  SureChEMBL SureChEMBL    / /  ZINC / ZINC /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | | OpenEye OEToolkits 1.5.0 | |
|---|
-PDB entries
Showing all 6 items

PDB-3e9z: 
Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with 6-chloroguanine
PDB-3fo4: 
Crystal structure of guanine riboswitch C74U mutant bound to 6-chloroguanine
PDB-3ger: 
Guanine riboswitch bound to 6-chloroguanine
PDB-3gog: 
Guanine riboswitch A21G,U75C mutant bound to 6-chloroguanine

PDB-6q62: 
Xanthomonas albilineans Dihydropteroate synthase in complex with (indole-2-carboxylic acid) and (6-chloroguanine)

PDB-6qoh: 
Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with Fragment 11 (2-Amino-6-chloropurine)
 Movie
Movie Controller
Controller



