[English] 日本語
 Yorodumi
Yorodumi- PDB-5u40: Human PPARdelta ligand-binding domain in complexed with specific ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5u40 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human PPARdelta ligand-binding domain in complexed with specific agonist 15 | |||||||||
 Components Components | Peroxisome proliferator-activated receptor delta | |||||||||
 Keywords Keywords | protein binding/activator / PPARdelta / ligand-binding domain / agonist / protein binding-activator complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationkeratinocyte migration / linoleic acid binding / positive regulation of skeletal muscle tissue regeneration / fat cell proliferation / positive regulation of epidermis development / axon ensheathment / positive regulation of fat cell proliferation / regulation of skeletal muscle satellite cell proliferation / D-glucose transmembrane transport / negative regulation of smooth muscle cell migration ...keratinocyte migration / linoleic acid binding / positive regulation of skeletal muscle tissue regeneration / fat cell proliferation / positive regulation of epidermis development / axon ensheathment / positive regulation of fat cell proliferation / regulation of skeletal muscle satellite cell proliferation / D-glucose transmembrane transport / negative regulation of smooth muscle cell migration / proteoglycan metabolic process / fatty acid catabolic process / negative regulation of collagen biosynthetic process / positive regulation of myoblast proliferation / positive regulation of fatty acid oxidation / phospholipid biosynthetic process / Carnitine shuttle / negative regulation of myoblast differentiation / response to vitamin A / Signaling by Retinoic Acid / positive regulation of fatty acid metabolic process / fatty acid beta-oxidation / cell-substrate adhesion / negative regulation of cholesterol storage / decidualization / nuclear steroid receptor activity / keratinocyte proliferation / NF-kappaB binding / adipose tissue development / positive regulation of fat cell differentiation / positive regulation of insulin secretion involved in cellular response to glucose stimulus / response to glucose / fatty acid transport / cellular response to nutrient levels / cholesterol metabolic process / energy homeostasis / intracellular receptor signaling pathway / hormone-mediated signaling pathway / embryo implantation / negative regulation of miRNA transcription / response to activity / generation of precursor metabolites and energy / apoptotic signaling pathway / negative regulation of smooth muscle cell proliferation / phosphatidylinositol 3-kinase/protein kinase B signal transduction / wound healing / fatty acid metabolic process / negative regulation of cell growth / Nuclear Receptor transcription pathway / lipid metabolic process / negative regulation of inflammatory response / DNA-binding transcription repressor activity, RNA polymerase II-specific / transcription coactivator binding / vasodilation / glucose metabolic process / nuclear receptor activity / negative regulation of epithelial cell proliferation / sequence-specific double-stranded DNA binding / heart development / cellular response to lipopolysaccharide / cellular response to hypoxia / DNA-binding transcription factor activity, RNA polymerase II-specific / cell differentiation / cell population proliferation / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of DNA-templated transcription / apoptotic process / lipid binding / positive regulation of gene expression / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / chromatin / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | |||||||||
 Authors Authors | Wu, C.-C. / Baiga, T.J. / Downes, M. / La Clair, J.J. / Atkins, A.R. / Richard, S.B. / Stockley-Noel, T.A. / Bowman, M.E. / Evans, R.M. / Noel, J.P. | |||||||||
 Citation Citation |  Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2017 Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2017Title: Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta. Authors: Wu, C.C. / Baiga, T.J. / Downes, M. / La Clair, J.J. / Atkins, A.R. / Richard, S.B. / Fan, W. / Stockley-Noel, T.A. / Bowman, M.E. / Noel, J.P. / Evans, R.M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5u40.cif.gz 5u40.cif.gz | 331.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5u40.ent.gz pdb5u40.ent.gz | 277.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5u40.json.gz 5u40.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5u40_validation.pdf.gz 5u40_validation.pdf.gz | 1.5 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5u40_full_validation.pdf.gz 5u40_full_validation.pdf.gz | 1.5 MB | Display | |
| Data in XML |  5u40_validation.xml.gz 5u40_validation.xml.gz | 22.2 KB | Display | |
| Data in CIF |  5u40_validation.cif.gz 5u40_validation.cif.gz | 31.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/u4/5u40 https://data.pdbj.org/pub/pdb/validation_reports/u4/5u40 ftp://data.pdbj.org/pub/pdb/validation_reports/u4/5u40 ftp://data.pdbj.org/pub/pdb/validation_reports/u4/5u40 | HTTPS FTP |
-Related structure data
| Related structure data |  5u3qC  5u3rC  5u3sC  5u3tC  5u3uC  5u3vC  5u3wC  5u3xC  5u3yC  5u3zC  5u41C  5u42C  5u43C  5u44C  5u45C  5u46C  1gwzS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 4 molecules AB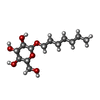

| #1: Protein | Mass: 31114.178 Da / Num. of mol.: 2 / Fragment: Ligand-binding domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PPARD, NR1C2, PPARB / Plasmid: pET28a(+) / Production host: Homo sapiens (human) / Gene: PPARD, NR1C2, PPARB / Plasmid: pET28a(+) / Production host:  #5: Sugar | |
|---|
-Non-polymers , 4 types, 193 molecules 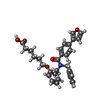





| #2: Chemical | | #3: Chemical | ChemComp-PEG / #4: Chemical | ChemComp-PGO / #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.85 Å3/Da / Density % sol: 56.79 % |
|---|---|
| Crystal grow | Temperature: 277.15 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: Bis-tris propane, potassium chloride, PEG 8000, 1,2-propandiol, EDTA, DTT PH range: 7.5 - 8.8 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.2.1 / Wavelength: 1 Å / Beamline: 8.2.1 / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Apr 13, 2006 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2→95.05 Å / Num. obs: 43342 / % possible obs: 91.9 % / Redundancy: 3 % / CC1/2: 0.975 / Net I/σ(I): 30.9 |
| Reflection shell | Resolution: 2→2.03 Å / Redundancy: 1.9 % / Mean I/σ(I) obs: 1 / CC1/2: 0.278 / % possible all: 61.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1GWZ Resolution: 2→95.05 Å / SU ML: 0.27 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 25.67
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→95.05 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




