Entry Database : PDB / ID : 5mazTitle STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PA14 IN COMPLEX WITH 3-Thiophenesulfonamide-2,5-dimethyl-N-methyl-beta-L-fucopyranoside Fucose-binding lectin PA-IIL Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / Biological species Pseudomonas aeruginosa (bacteria)Method / / / Resolution : 1.45 Å Authors Sommer, R. / Imberty, A. / Titz, A. / Varrot, A. Funding support 1items Organization Grant number Country COST COST-STSM-ECOST-STSM-BM1003-240314-042549
Journal : To Be Published Title : Discovery of a new class of C-glycoside based inhibitors of the virulence factor LecB from Pseudomonas aeruginosaAuthors : Sommer, R. / Wagner, S. / Varrot, A. / Hauk, D. / Ryckmans, T. / Hartman, R.W. / Brunner, T. / Rademacher, C. / Imberty, A. / Titz, A. History Deposition Nov 7, 2016 Deposition site / Processing site Revision 1.0 Dec 20, 2017 Provider / Type Revision 1.1 Jul 29, 2020 Group / Derived calculations / Structure summaryCategory chem_comp / entity ... chem_comp / entity / pdbx_chem_comp_identifier / pdbx_entity_nonpoly / pdbx_struct_conn_angle / struct_conn / struct_conn_type / struct_site / struct_site_gen Item _chem_comp.name / _chem_comp.type ... _chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_entity_nonpoly.name / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn_type.id Description / Provider / Type Revision 1.2 Jan 17, 2024 Group Data collection / Database references ... Data collection / Database references / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim Item _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI ... _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.45 Å
MOLECULAR REPLACEMENT / Resolution: 1.45 Å  Authors
Authors Citation
Citation Journal: To Be Published
Journal: To Be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5maz.cif.gz
5maz.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5maz.ent.gz
pdb5maz.ent.gz PDB format
PDB format 5maz.json.gz
5maz.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 5maz_validation.pdf.gz
5maz_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 5maz_full_validation.pdf.gz
5maz_full_validation.pdf.gz 5maz_validation.xml.gz
5maz_validation.xml.gz 5maz_validation.cif.gz
5maz_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/ma/5maz
https://data.pdbj.org/pub/pdb/validation_reports/ma/5maz ftp://data.pdbj.org/pub/pdb/validation_reports/ma/5maz
ftp://data.pdbj.org/pub/pdb/validation_reports/ma/5maz
 Links
Links Assembly
Assembly
 Components
Components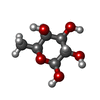

 Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)
Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)

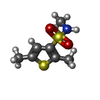







 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID23-2 / Wavelength: 0.8726 Å
/ Beamline: ID23-2 / Wavelength: 0.8726 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj



