[English] 日本語
 Yorodumi
Yorodumi- EMDB-5822: Negative-stain electron microscopy reconstruction of Tetrahymena ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5822 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
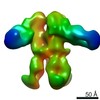
| Title | Negative-stain electron microscopy reconstruction of Tetrahymena telomerase (Teb1-f, affinity labeled by anti-FLAG Fab) | |||||||||
 Map data Map data | 3D reconstruction of Tetrahymena telomerase (Teb1-f, affinity labeled by anti-FLAG Fab) using Random Conical Tilt (RCT) method | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | telomerase / telomere / Tetrahymena | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 30.0 Å | |||||||||
 Authors Authors | Jiang J / Miracco EJ / Hong K / Eckert B / Chan H / Cash DD / Min B / Zhou ZH / Collins K / Feigon J | |||||||||
 Citation Citation |  Journal: Nature / Year: 2013 Journal: Nature / Year: 2013Title: The architecture of Tetrahymena telomerase holoenzyme. Authors: Jiansen Jiang / Edward J Miracco / Kyungah Hong / Barbara Eckert / Henry Chan / Darian D Cash / Bosun Min / Z Hong Zhou / Kathleen Collins / Juli Feigon /  Abstract: Telomerase adds telomeric repeats to chromosome ends using an internal RNA template and a specialized telomerase reverse transcriptase (TERT), thereby maintaining genome integrity. Little is known ...Telomerase adds telomeric repeats to chromosome ends using an internal RNA template and a specialized telomerase reverse transcriptase (TERT), thereby maintaining genome integrity. Little is known about the physical relationships among protein and RNA subunits within a biologically functional holoenzyme. Here we describe the architecture of Tetrahymena thermophila telomerase holoenzyme determined by electron microscopy. Six of the seven proteins and the TERT-binding regions of telomerase RNA (TER) have been localized by affinity labelling. Fitting with high-resolution structures reveals the organization of TERT, TER and p65 in the ribonucleoprotein (RNP) catalytic core. p50 has an unanticipated role as a hub between the RNP catalytic core, p75-p19-p45 subcomplex, and the DNA-binding Teb1. A complete in vitro holoenzyme reconstitution assigns function to these interactions in processive telomeric repeat synthesis. These studies provide the first view of the extensive network of subunit associations necessary for telomerase holoenzyme assembly and physiological function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5822.map.gz emd_5822.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5822-v30.xml emd-5822-v30.xml emd-5822.xml emd-5822.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5822.png emd_5822.png | 63.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5822 http://ftp.pdbj.org/pub/emdb/structures/EMD-5822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5822 | HTTPS FTP |
-Validation report
| Summary document |  emd_5822_validation.pdf.gz emd_5822_validation.pdf.gz | 78.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5822_full_validation.pdf.gz emd_5822_full_validation.pdf.gz | 77.6 KB | Display | |
| Data in XML |  emd_5822_validation.xml.gz emd_5822_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5822 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5822 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5822 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5822 | HTTPS FTP |
-Related structure data
| Related structure data |  5804C  5807C  5808C  5809C  5810C  5811C  5812C  5813C  5814C  5815C  5816C  5817C  5818C  5819C  5820C  5821C 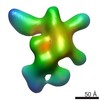 5823C  5824C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5822.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5822.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of Tetrahymena telomerase (Teb1-f, affinity labeled by anti-FLAG Fab) using Random Conical Tilt (RCT) method | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.41 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Telomerase holoenzyme (Teb1-f, labeled by anti-FLAG Fab) from Tet...
| Entire | Name: Telomerase holoenzyme (Teb1-f, labeled by anti-FLAG Fab) from Tetrahymena thermophila |
|---|---|
| Components |
|
-Supramolecule #1000: Telomerase holoenzyme (Teb1-f, labeled by anti-FLAG Fab) from Tet...
| Supramolecule | Name: Telomerase holoenzyme (Teb1-f, labeled by anti-FLAG Fab) from Tetrahymena thermophila type: sample / ID: 1000 / Oligomeric state: Hetero-octamer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Telomerase holoenzyme
| Macromolecule | Name: Telomerase holoenzyme / type: protein_or_peptide / ID: 1 / Name.synonym: Telomerase / Number of copies: 1 / Oligomeric state: Hetero-octamer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #2: Fab from monoclonal anit-FLAG M2 antibody
| Macromolecule | Name: Fab from monoclonal anit-FLAG M2 antibody / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: monomer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 20 mM HEPES, 50 mM NaCl, 1 mM MgCl2, 1 mM TCEP |
|---|---|
| Staining | Type: NEGATIVE / Details: 0.8% uranyl formate |
| Grid | Details: 200 mesh grid with thin carbon support |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 298 K |
| Date | May 18, 2012 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Number real images: 339 / Average electron dose: 40 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 68027 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.7 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 70000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle min: 65 / Tilt angle max: 65 |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The RCT 3D reconstruction was done using SPIDER. |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: SPIDER / Number images used: 654 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















