[English] 日本語
 Yorodumi
Yorodumi- EMDB-4734: Structure of the core Shigella flexneri type III secretion system... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4734 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the core Shigella flexneri type III secretion system export gate complex SctRST (Spa24/Spa9/Spa29). | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Type III secretion / T3SS / Flagella / Cryo-EM / Membrane protein / Protein transport | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  Shigella flexneri (bacteria) Shigella flexneri (bacteria) | ||||||||||||||||||
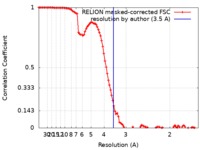
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||
 Authors Authors | Johnson S / Kuhlen L | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structure of the core of the type III secretion system export apparatus. Authors: Lucas Kuhlen / Patrizia Abrusci / Steven Johnson / Joseph Gault / Justin Deme / Joseph Caesar / Tobias Dietsche / Mehari Tesfazgi Mebrhatu / Tariq Ganief / Boris Macek / Samuel Wagner / ...Authors: Lucas Kuhlen / Patrizia Abrusci / Steven Johnson / Joseph Gault / Justin Deme / Joseph Caesar / Tobias Dietsche / Mehari Tesfazgi Mebrhatu / Tariq Ganief / Boris Macek / Samuel Wagner / Carol V Robinson / Susan M Lea /   Abstract: Export of proteins through type III secretion systems is critical for motility and virulence of many major bacterial pathogens. Three putative integral membrane proteins (FliP, FliQ, FliR) are ...Export of proteins through type III secretion systems is critical for motility and virulence of many major bacterial pathogens. Three putative integral membrane proteins (FliP, FliQ, FliR) are suggested to form the core of an export gate in the inner membrane, but their structure, assembly and location within the final nanomachine remain unclear. Here, we present the cryoelectron microscopy structure of the Salmonella Typhimurium FliP-FliQ-FliR complex at 4.2 Å. None of the subunits adopt canonical integral membrane protein topologies, and common helix-turn-helix structural elements allow them to form a helical assembly with 5:4:1 stoichiometry. Fitting of the structure into reconstructions of intact secretion systems, combined with cross-linking, localize the export gate as a core component of the periplasmic portion of the machinery. This study thereby identifies the export gate as a key element of the secretion channel and implies that it primes the helical architecture of the components assembling downstream. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4734.map.gz emd_4734.map.gz | 70.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4734-v30.xml emd-4734-v30.xml emd-4734.xml emd-4734.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4734_fsc.xml emd_4734_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4734.png emd_4734.png | 59.8 KB | ||
| Masks |  emd_4734_msk_1.map emd_4734_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4734.cif.gz emd-4734.cif.gz | 6.5 KB | ||
| Others |  emd_4734_additional.map.gz emd_4734_additional.map.gz emd_4734_half_map_1.map.gz emd_4734_half_map_1.map.gz emd_4734_half_map_2.map.gz emd_4734_half_map_2.map.gz | 6.8 MB 71.3 MB 71.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4734 http://ftp.pdbj.org/pub/emdb/structures/EMD-4734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4734 | HTTPS FTP |
-Related structure data
| Related structure data |  6r6bMC  4733C  6r69C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4734.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4734.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4734_msk_1.map emd_4734_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_4734_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: None
| File | emd_4734_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: None
| File | emd_4734_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Shigella flexneri type III secretion export gate (Spa24Spa9Spa29)
| Entire | Name: Shigella flexneri type III secretion export gate (Spa24Spa9Spa29) |
|---|---|
| Components |
|
-Supramolecule #1: Shigella flexneri type III secretion export gate (Spa24Spa9Spa29)
| Supramolecule | Name: Shigella flexneri type III secretion export gate (Spa24Spa9Spa29) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 191 KDa |
-Macromolecule #1: Surface presentation of antigens protein SpaP
| Macromolecule | Name: Surface presentation of antigens protein SpaP / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 24.215562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLSDMSLIAT LSFFTLLPFL VAAGTCYIKF SIVFVMVRNA LGLQQVPSNM TLNGIALIMA LFVMKPIIEA GYENYLNGPQ KFDTISDIV RFSDSGLMEY KQYLKKHTDL ELARFFQRSE EENADLKSAE NNDYSLFSLL PAYALSEIKD AFKIGFYLYL P FVVVDLVI ...String: MLSDMSLIAT LSFFTLLPFL VAAGTCYIKF SIVFVMVRNA LGLQQVPSNM TLNGIALIMA LFVMKPIIEA GYENYLNGPQ KFDTISDIV RFSDSGLMEY KQYLKKHTDL ELARFFQRSE EENADLKSAE NNDYSLFSLL PAYALSEIKD AFKIGFYLYL P FVVVDLVI SSILLALGMM MMSPITISVP IKLVLFVALD GWGILSKALI EQYINIPA UniProtKB: Surface presentation of antigens protein SpaP |
-Macromolecule #2: Surface presentation of antigens protein SpaR
| Macromolecule | Name: Surface presentation of antigens protein SpaR / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 32.644086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDISSWFESI HVFLILLNGV FFRLAPLFFF LPFLNNGIIS PSIRIPVIFL VASGLITSGK VDIGSSVFEH VYFLMFKEII VGLLLSFCL SLPFWIFHAV GSIIDNQRGA TLSSSIDPAN GVDTSELAKF FNLFSAVVFL YSGGMVFILE SIQLSYNICP L FSQCSFRI ...String: MDISSWFESI HVFLILLNGV FFRLAPLFFF LPFLNNGIIS PSIRIPVIFL VASGLITSGK VDIGSSVFEH VYFLMFKEII VGLLLSFCL SLPFWIFHAV GSIIDNQRGA TLSSSIDPAN GVDTSELAKF FNLFSAVVFL YSGGMVFILE SIQLSYNICP L FSQCSFRI SNILTFLTLL ASQAVILASP VMIVLLLSEV LLGVLSRFAP QMNAFSVSLT IKSLLAIFII FICSSTIYFS KV QFFLGEH KFFTNLFVRE NLYFQGQFGS WSHPQFEKGG GSGGGSGGGS WSHPQFEK UniProtKB: Surface presentation of antigens protein SpaR |
-Macromolecule #3: Surface presentation of antigens protein SpaQ
| Macromolecule | Name: Surface presentation of antigens protein SpaQ / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 9.433338 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDIVYMGNK ALYLILIFSL WPVGIATVIG LSIGLLQTVT QLQEQTLPFG IKLIGVSISL LLLSGWYGEV LLSFCHEIMF LIKSGV UniProtKB: Surface presentation of antigens protein SpaQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.4 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 3741 / Average exposure time: 8.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6r6b: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)