+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4616 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis of cohesin ring opening | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chromatin / genome segregation / cohesin / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationEstablishment of Sister Chromatid Cohesion / Resolution of Sister Chromatid Cohesion / meiotic cohesin complex / establishment of meiotic sister chromatid cohesion / cohesin complex / mitotic cohesin complex / synaptonemal complex assembly / SUMOylation of DNA damage response and repair proteins / meiotic sister chromatid cohesion / establishment of mitotic sister chromatid cohesion ...Establishment of Sister Chromatid Cohesion / Resolution of Sister Chromatid Cohesion / meiotic cohesin complex / establishment of meiotic sister chromatid cohesion / cohesin complex / mitotic cohesin complex / synaptonemal complex assembly / SUMOylation of DNA damage response and repair proteins / meiotic sister chromatid cohesion / establishment of mitotic sister chromatid cohesion / replication-born double-strand break repair via sister chromatid exchange / mitotic chromosome condensation / reciprocal meiotic recombination / mitotic sister chromatid cohesion / sister chromatid cohesion / protein acetylation / chromosome, centromeric region / condensed nuclear chromosome / G2/M transition of mitotic cell cycle / double-strand break repair / double-stranded DNA binding / cell division / apoptotic process / DNA damage response / chromatin binding / protein kinase binding / ATP hydrolysis activity / mitochondrion / DNA binding / ATP binding / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) / Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) /   Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) / Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) /  | |||||||||
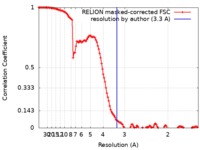
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Panne D / Muir KW | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening. Authors: Kyle W Muir / Yan Li / Felix Weis / Daniel Panne /    Abstract: Genome regulation requires control of chromosome organization by SMC-kleisin complexes. The cohesin complex contains the Smc1 and Smc3 subunits that associate with the kleisin Scc1 to form a ring- ...Genome regulation requires control of chromosome organization by SMC-kleisin complexes. The cohesin complex contains the Smc1 and Smc3 subunits that associate with the kleisin Scc1 to form a ring-shaped complex that can topologically engage chromatin to regulate chromatin structure. Release from chromatin involves opening of the ring at the Smc3-Scc1 interface in a reaction that is controlled by acetylation and engagement of the Smc ATPase head domains. To understand the underlying molecular mechanisms, we have determined the 3.2-Å resolution cryo-electron microscopy structure of the ATPγS-bound, heterotrimeric cohesin ATPase head module and the 2.1-Å resolution crystal structure of a nucleotide-free Smc1-Scc1 subcomplex from Saccharomyces cerevisiae and Chaetomium thermophilium. We found that ATP-binding and Smc1-Smc3 heterodimerization promote conformational changes within the ATPase that are transmitted to the Smc coiled-coil domains. Remodeling of the coiled-coil domain of Smc3 abrogates the binding surface for Scc1, thus leading to ring opening at the Smc3-Scc1 interface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4616.map.gz emd_4616.map.gz | 9.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4616-v30.xml emd-4616-v30.xml emd-4616.xml emd-4616.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4616_fsc.xml emd_4616_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_4616.png emd_4616.png | 53 KB | ||
| Filedesc metadata |  emd-4616.cif.gz emd-4616.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4616 http://ftp.pdbj.org/pub/emdb/structures/EMD-4616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4616 | HTTPS FTP |
-Related structure data
| Related structure data |  6qpwMC  6qpqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4616.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4616.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cohesin ATPase head module
| Entire | Name: Cohesin ATPase head module |
|---|---|
| Components |
|
-Supramolecule #1: Cohesin ATPase head module
| Supramolecule | Name: Cohesin ATPase head module / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: Structural maintenance of chromosomes protein,Structural maintena...
| Supramolecule | Name: Structural maintenance of chromosomes protein,Structural maintenance of chromosomes protein type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) |
-Supramolecule #3: Sister chromatid cohesion protein 1, Structural maintenance of ch...
| Supramolecule | Name: Sister chromatid cohesion protein 1, Structural maintenance of chromosomes protein 3,Structural maintenance of chromosomes protein 3 type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Structural maintenance of chromosomes protein,Structural maintena...
| Macromolecule | Name: Structural maintenance of chromosomes protein,Structural maintenance of chromosomes protein type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) |
| Molecular weight | Theoretical: 27.570475 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKLIRLELF NFKSYKGHHT LLFGDSYFTS IIGPNGSGKS NSMDAISFVL GIKSSHLRSS NLRDLIYRGR VMKTSKIQDD GTTAPATNG DVNGYENGDA GDDEDTSQRT SRNDPKTAWV MAVYEDDAGE LHRWKRTITA NGTSEYRIND RVVNAQQYNE A LEKENILI ...String: MGKLIRLELF NFKSYKGHHT LLFGDSYFTS IIGPNGSGKS NSMDAISFVL GIKSSHLRSS NLRDLIYRGR VMKTSKIQDD GTTAPATNG DVNGYENGDA GDDEDTSQRT SRNDPKTAWV MAVYEDDAGE LHRWKRTITA NGTSEYRIND RVVNAQQYNE A LEKENILI KARNFLVFQG DVEAIASQSP QDLTRLIEQI SGSLEYKEEY ERLEEEVRQA TEEQAYKLQR RRAANSEIKQ YM EQ UniProtKB: Structural maintenance of chromosomes protein, Structural maintenance of chromosomes protein |
-Macromolecule #2: Sister chromatid cohesion protein 1
| Macromolecule | Name: Sister chromatid cohesion protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.48994 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASKAIVQMA KILRKELSEE KEVIFTDVLK SQANTEPENI TKREASRGFF DILSLATEGC IGLSQTEAFG NIKIDAKPAL FERFI UniProtKB: Sister chromatid cohesion protein 1 |
-Macromolecule #3: Structural maintenance of chromosomes protein 3,Structural mainte...
| Macromolecule | Name: Structural maintenance of chromosomes protein 3,Structural maintenance of chromosomes protein 3 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.051789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAYIKRVIIK GFKTYRNETI IDNFSPHQNV IIGSNGSGKS NFFAAIRFVL SDDYSNLKRE ERQGLIHQGS GGSVMSASVE IVFHDPDHS MILPSGVLSR GDDEVTIRRT VGLKKDDYQL NDRNVTKGDI VRMLETAGFS MNNPYNIVPQ GKIVALTNAK D KERLQLLE ...String: MAYIKRVIIK GFKTYRNETI IDNFSPHQNV IIGSNGSGKS NFFAAIRFVL SDDYSNLKRE ERQGLIHQGS GGSVMSASVE IVFHDPDHS MILPSGVLSR GDDEVTIRRT VGLKKDDYQL NDRNVTKGDI VRMLETAGFS MNNPYNIVPQ GKIVALTNAK D KERLQLLE DVVGAKSFEV KLKASLKKME ETEQKKIQIN KEMGELNSKL SEMEQERKEL EKYNELERNR KIYQFTLYDR EL NEVINQM ERLDGDYNNT VYSSESSKHP TSLVPRGSDI TSDQLLQRLN DMNTEISGLK NVNKRAFENF KKFNERRKDL AER ASELDE SKDSIQDLIV KLKQQKVNAV DSTFQKVSEN FEAVFERLVP RGTAKLIIHR KNDNANDHDE SIDVDMDAES NESQ NGKDS EIMYTGVSIS VSFNSKQNEQ LHVEQLSGGQ KTVCAIALIL AIQMVDPASF YLFDEIDACL DKQYRTAVAT LLKEL SKNA QFICTTFRTD MLQVADKFFR VKYECKISTV IEVNREEAIG FIRGSNKFAE V UniProtKB: Structural maintenance of chromosomes protein 3, Structural maintenance of chromosomes protein 3 |
-Macromolecule #4: Sister chromatid cohesion protein 1,Structural maintenance of chr...
| Macromolecule | Name: Sister chromatid cohesion protein 1,Structural maintenance of chromosomes protein type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) Chaetomium thermophilum var. thermophilum DSM 1495 (fungus) |
| Molecular weight | Theoretical: 42.292125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVTENPQRLT VLRLATNKGP LAQIWLASNM SNIPRGSVIQ THIAESAKEI AKASGSDDES GDNEYITLRT SGELLQGIVR VYSKQATFL LTDIKDTLTK ISMLFKTSQK MTSTVNRLNT VTRVHQLMLE DAVTEREVLV TPGLEFLDDT TIPVGLMAQE N PNLRAMDR ...String: MVTENPQRLT VLRLATNKGP LAQIWLASNM SNIPRGSVIQ THIAESAKEI AKASGSDDES GDNEYITLRT SGELLQGIVR VYSKQATFL LTDIKDTLTK ISMLFKTSQK MTSTVNRLNT VTRVHQLMLE DAVTEREVLV TPGLEFLDDT TIPVGLMAQE N PNLRAMDR LDHVRKQLEQ TEQEFEASKA KLRQARESFQ AVKQKRLELF NKAFTHIQEQ ITHVYKELTR SEAYPLGGQA YL DIEEDTD TPFLSGVKYH AMPPCKRFRD MEHLSGGEKT MAALALLFAI HSYQPSPFFV LDEVDCALDN ANVEKIKKYI REH AGPGMQ FIVISLKPAL FQASESLIGV YRDQEANTSR TLTLDLRKYR HHHHHH UniProtKB: Sister chromatid cohesion protein 1, Structural maintenance of chromosomes protein |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 6 / Number of copies: 2 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 42.08 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)