+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mechanistic Insights Revealed by YbtPQ in the Occluded State | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC importer / siderophore / occluded / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances / ATPase-coupled lipid transmembrane transporter activity / ABC-type transporter activity / ATP hydrolysis activity / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Hu W / Parkinson C / Zheng H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biomolecules / Year: 2024 Journal: Biomolecules / Year: 2024Title: Mechanistic Insights Revealed by YbtPQ in the Occluded State. Authors: Wenxin Hu / Chance Parkinson / Hongjin Zheng /  Abstract: Recently, several ATP-binding cassette (ABC) importers have been found to adopt the typical fold of type IV ABC exporters. Presumably, these importers would function under the transport scheme of ...Recently, several ATP-binding cassette (ABC) importers have been found to adopt the typical fold of type IV ABC exporters. Presumably, these importers would function under the transport scheme of "alternating access" like those exporters, cycling through inward-open, occluded, and outward-open conformations. Understanding how the exporter-like importers move substrates in the opposite direction requires structural studies on all the major conformations. To shed light on this, here we report the structure of yersiniabactin importer YbtPQ from uropathogenic in the occluded conformation trapped by ADP-vanadate (ADP-Vi) at a 3.1 Å resolution determined by cryo-electron microscopy. The structure shows unusual local rearrangements in multiple helices and loops in its transmembrane domains (TMDs). In addition, the dimerization of the nucleotide-binding domains (NBDs) promoted by the vanadate trapping is highlighted by the "screwdriver" action at one of the two hinge points. These structural observations are rare and thus provide valuable information to understand the structural plasticity of the exporter-like ABC importers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43498.map.gz emd_43498.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43498-v30.xml emd-43498-v30.xml emd-43498.xml emd-43498.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43498.png emd_43498.png | 93 KB | ||
| Filedesc metadata |  emd-43498.cif.gz emd-43498.cif.gz | 6.2 KB | ||
| Others |  emd_43498_half_map_1.map.gz emd_43498_half_map_1.map.gz emd_43498_half_map_2.map.gz emd_43498_half_map_2.map.gz | 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43498 http://ftp.pdbj.org/pub/emdb/structures/EMD-43498 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43498 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43498 | HTTPS FTP |
-Validation report
| Summary document |  emd_43498_validation.pdf.gz emd_43498_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43498_full_validation.pdf.gz emd_43498_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_43498_validation.xml.gz emd_43498_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_43498_validation.cif.gz emd_43498_validation.cif.gz | 13.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43498 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43498 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43498 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43498 | HTTPS FTP |
-Related structure data
| Related structure data |  8vsiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43498.map.gz / Format: CCP4 / Size: 5.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43498.map.gz / Format: CCP4 / Size: 5.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_43498_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
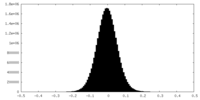
| Density Histograms |
-Half map: #2
| File | emd_43498_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
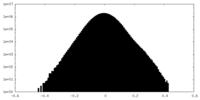
| Density Histograms |
- Sample components
Sample components
-Entire : YbtPQ
| Entire | Name: YbtPQ |
|---|---|
| Components |
|
-Supramolecule #1: YbtPQ
| Supramolecule | Name: YbtPQ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ABC transporter ATP-binding protein
| Macromolecule | Name: ABC transporter ATP-binding protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.348664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSQSSNTES LSRFPLWQVI TPVRRKVILA MALAGLAALT SLGALLFLAW SLRDIRATPD AIPAWPLGGV IGCVVLTFVL RLQAFNTSH YAAFHLENIL RSRLARKALQ LPPGVLQQMG SGSVAKVMLD DVKSLHIFVA DSTPLYARAI IMPLATIVIL F WLDWRLAI ...String: MSSQSSNTES LSRFPLWQVI TPVRRKVILA MALAGLAALT SLGALLFLAW SLRDIRATPD AIPAWPLGGV IGCVVLTFVL RLQAFNTSH YAAFHLENIL RSRLARKALQ LPPGVLQQMG SGSVAKVMLD DVKSLHIFVA DSTPLYARAI IMPLATIVIL F WLDWRLAI ATLGVLAFGS VVLVLARQRS ENMAQRYHKA REQVSAAVIE FVQAMPVVRT FDSGSTSFLR YQRALEEWVD VL KTWYRKA GFSARFSFSI LNPLPTLFVL IWSGYGLLHY GSFDFIAWVA VLLLGSGMAE AVMPMMMLNN LVAQTRLSIQ RIY QVLAMP ELSLPQSDQQ PQEASITFEQ VSFHYPQART GAALQEVSFH VPAGQIVALV GPSGAGKSTV ARLLLRYADP DKGH IRIGG VDLRDMQTDT LMKQLSFVFQ DNFLFADTIA NNIRLGAPDT PLEAVIAAAR VAQAHDFISA LPEGYNTRVG ERGVF LSGG QRQRITIARA LLQDRPILVL DEATAFADPE NEAALIKALA AAMRGRTVIM VAHRLSMVTQ ADVILLFSDG QLREMG NHT QLLAQGGLYQ RLWQHYQQAQ HWVPGGTQEE VVENERQ UniProtKB: ABC transporter ATP-binding protein |
-Macromolecule #2: Permease and ATP-binding protein of yersiniabactin-iron ABC trans...
| Macromolecule | Name: Permease and ATP-binding protein of yersiniabactin-iron ABC transporter YbtQ type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.526086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKDNNPADNL AWRVIWRQLI SSVGSQARML RRSMLALLLA AFMQGIAFAC LYPIIDALLR GDAPQLLNWA MAFSVAAIVT LVLRWYGLG FEYRGHLAQA THELRLRLGE QLRRVPLEKL QRGRAGEMNA LLLGSVDENL NYVIAIANIL LLTIVTPLTA S LATLWIDW ...String: MKDNNPADNL AWRVIWRQLI SSVGSQARML RRSMLALLLA AFMQGIAFAC LYPIIDALLR GDAPQLLNWA MAFSVAAIVT LVLRWYGLG FEYRGHLAQA THELRLRLGE QLRRVPLEKL QRGRAGEMNA LLLGSVDENL NYVIAIANIL LLTIVTPLTA S LATLWIDW RLGLVMLLIF PLLVPFYYWR RPAMRRQMQT LGEAHQRLSG DIVEFAQGMM VLRTCGSDAD KSRALLAHFN AL ENLQTRT HRQGAGATML IASVVELGLQ VVVLSGIVWV VTGTLNLAFL IAAVAMIMRF AEPMAMFISY TSVVELIASA LQR IEQFMA IAPLPVAEQS EMPERYDIRF DNVSYRYEEG DGYALNHVSL TFPAASMSAL VGASGAGKTT VTKLLMRYAD PQQG QISIG GVDIRRLTPE QLNSLISVVF QDVWLFDDTL LANIRIARPQ ATRQEVEEAA RAAQCLEFIS RLPQGWLTPM GEMGG QLSG GERQRISIAR ALLKNAPVVI LDEPTAALDI ESELAVQKAI DNLVHNRTVI IIAHRLSTIA GAGNILVMEE GQVVEQ GTH AQLLSHHGRY QALWQAQMAA RVWRDDGVSA SGEWVHE UniProtKB: Permease and ATP-binding protein of yersiniabactin-iron ABC transporter YbtQ |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ADP ORTHOVANADATE
| Macromolecule | Name: ADP ORTHOVANADATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: AOV |
|---|---|
| Molecular weight | Theoretical: 544.156 Da |
| Chemical component information | 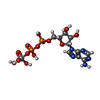 ChemComp-AOV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 340080 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)