+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM Structure of Maize Streak Virus (MSV) - Geminivirus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample | Maize streak virus - [Nigeria] != Maize streak virus genotype A (isolate Nigeria) Maize streak virus - [Nigeria]
| |||||||||
 Keywords Keywords | Plant / ssDNA / Icosahedron / capsid / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=1 icosahedral viral capsid / viral penetration into host nucleus / host cell / symbiont entry into host cell / host cell nucleus / structural molecule activity / DNA binding Similarity search - Function | |||||||||
| Biological species |  Maize streak virus genotype A (isolate Nigeria) Maize streak virus genotype A (isolate Nigeria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | McKenna R / Bennett AB / Mietzsch M / Hull JA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The two states of Maize Streak Virus (MSV) Geminivirus Architecture Authors: McKenna R / Bennett AB / Mietzsch M / Hull JA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42232.map.gz emd_42232.map.gz | 589.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42232-v30.xml emd-42232-v30.xml emd-42232.xml emd-42232.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42232.png emd_42232.png | 35.3 KB | ||
| Filedesc metadata |  emd-42232.cif.gz emd-42232.cif.gz | 5.8 KB | ||
| Others |  emd_42232_half_map_1.map.gz emd_42232_half_map_1.map.gz emd_42232_half_map_2.map.gz emd_42232_half_map_2.map.gz | 379 MB 379 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42232 http://ftp.pdbj.org/pub/emdb/structures/EMD-42232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42232 | HTTPS FTP |
-Validation report
| Summary document |  emd_42232_validation.pdf.gz emd_42232_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42232_full_validation.pdf.gz emd_42232_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_42232_validation.xml.gz emd_42232_validation.xml.gz | 19.7 KB | Display | |
| Data in CIF |  emd_42232_validation.cif.gz emd_42232_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42232 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42232 | HTTPS FTP |
-Related structure data
| Related structure data |  8ugqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42232.map.gz / Format: CCP4 / Size: 634.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42232.map.gz / Format: CCP4 / Size: 634.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_42232_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
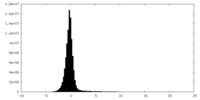
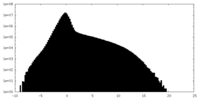
| Density Histograms |
-Half map: #2
| File | emd_42232_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
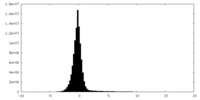
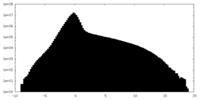
| Density Histograms |
- Sample components
Sample components
-Entire : Maize streak virus - [Nigeria]
| Entire | Name: Maize streak virus - [Nigeria] |
|---|---|
| Components |
|
-Supramolecule #1: Maize streak virus genotype A (isolate Nigeria)
| Supramolecule | Name: Maize streak virus genotype A (isolate Nigeria) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Gemini capsid is a Pseudo T=1 icosahedron. The structure contains both protein and ssDNA. NCBI-ID: 10823 Sci species name: Maize streak virus genotype A (isolate Nigeria) Sci species strain: isolate Nigeria / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Maize streak virus genotype A (isolate Nigeria) / Strain: isolate Nigeria Maize streak virus genotype A (isolate Nigeria) / Strain: isolate Nigeria |
| Molecular weight | Theoretical: 26.880529 KDa |
| Recombinant expression | Organism:  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) |
| Sequence | String: MSTSKRKRGD DSNWSKRVTK KKPSSAGLKR AGSKADRPSL QIQTLQHAGT TMITVPSGGV CDLINTYARG SDEGNRHTSE TLTYKIAID YHFVADAAAC RYSNTGTGVM WLVYDTTPGG QAPTPQTIFA YPDTLKAWPA TWKVSRELCH RFVVKRRWLF N METDGRIG ...String: MSTSKRKRGD DSNWSKRVTK KKPSSAGLKR AGSKADRPSL QIQTLQHAGT TMITVPSGGV CDLINTYARG SDEGNRHTSE TLTYKIAID YHFVADAAAC RYSNTGTGVM WLVYDTTPGG QAPTPQTIFA YPDTLKAWPA TWKVSRELCH RFVVKRRWLF N METDGRIG SDIPPSNASW KPCKRNIYFH KFTSGLGVRT QWKNVTDGGV GAIQRGALYM VIAPGNGLTF TAHGQTRLYF KS VGN UniProtKB: Capsid protein |
-Macromolecule #2: DNA (5'-D(P*CP*GP*AP*AP*CP*CP*CP*CP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*AP*AP*CP*CP*CP*CP*A)-3') / type: dna / ID: 2 / Number of copies: 11 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Maize streak virus genotype A (isolate Nigeria) / Strain: isolate Nigeria Maize streak virus genotype A (isolate Nigeria) / Strain: isolate Nigeria |
| Molecular weight | Theoretical: 2.669778 KDa |
| Sequence | String: (DC)(DG)(DA)(DA)(DC)(DC)(DC)(DC)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 4 / Component - Concentration: 0.1 M / Component - Formula: NaAc / Component - Name: Sodium acetate |
| Grid | Model: C-flat / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-64 (8k x 8k) / Number real images: 1408 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.6270000000000002 µm / Nominal defocus min: 0.9470000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D5 (2x5 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 3.17 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 1408 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
-Atomic model buiding 1
| Initial model | Chain - Initial model type: in silico model |
|---|---|
| Refinement | Protocol: AB INITIO MODEL |
| Output model |  PDB-8ugq: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)