+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Capsid of Rat Bocavirus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Parvovirus / RBoV / Rat Bocavirus / Capsid / VIRUS | |||||||||
| Function / homology | Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / VP2 Function and homology information Function and homology information | |||||||||
| Biological species |  Rat bocavirus Rat bocavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.52 Å | |||||||||
 Authors Authors | Velez M / Mietzsch M / McKenna R / Afione S / Zeher A / Huang R / Chiorini J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Viruses / Year: 2023 Journal: Viruses / Year: 2023Title: Structural Characterization of Canine Minute Virus, Rat and Porcine Bocavirus. Authors: Michael Velez / Mario Mietzsch / Jane Hsi / Logan Bell / Paul Chipman / Xiaofeng Fu / Robert McKenna /  Abstract: is an expansive genus of the , with a wide range of vertebrate hosts. This study investigates Canine minute virus (CnMV), Rat bocavirus (RBoV), and Porcine bocavirus 1 (PBoV1). Both CnMV and PBoV1 ... is an expansive genus of the , with a wide range of vertebrate hosts. This study investigates Canine minute virus (CnMV), Rat bocavirus (RBoV), and Porcine bocavirus 1 (PBoV1). Both CnMV and PBoV1 have been found in gastrointestinal infections in their respective hosts, with CnMV responsible for spontaneous abortions in dogs, while PBoV has been associated with encephalomyelitis in piglets. The pathogenicity of the recently identified RBoV is currently unknown. To initiate the characterization of these viruses, their capsids structures were determined by cryo-electron microscopy at resolutions ranging from 2.3 to 2.7 Å. Compared to other parvoviruses, the CnMV, PBoV1, and RBoV capsids showed conserved features, such as the channel at the fivefold symmetry axis. However, major differences were observed at the two- and threefold axes. While CnMV displays prominent threefold protrusions, the same region is more recessed in PBoV1 and RBoV. Furthermore, the typical twofold axis depression of parvoviral capsids is absent in CnMV or very small in PBoV and RBoV. These capsid structures extend the structural portfolio for the genus and will allow future characterization of these pathogens on a molecular level. This is important, as no antivirals or vaccines exist for these viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41616.map.gz emd_41616.map.gz | 263.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41616-v30.xml emd-41616-v30.xml emd-41616.xml emd-41616.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41616.png emd_41616.png | 272.8 KB | ||
| Filedesc metadata |  emd-41616.cif.gz emd-41616.cif.gz | 5.4 KB | ||
| Others |  emd_41616_half_map_1.map.gz emd_41616_half_map_1.map.gz emd_41616_half_map_2.map.gz emd_41616_half_map_2.map.gz | 110.9 MB 110.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41616 http://ftp.pdbj.org/pub/emdb/structures/EMD-41616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41616 | HTTPS FTP |
-Validation report
| Summary document |  emd_41616_validation.pdf.gz emd_41616_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41616_full_validation.pdf.gz emd_41616_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_41616_validation.xml.gz emd_41616_validation.xml.gz | 17 KB | Display | |
| Data in CIF |  emd_41616_validation.cif.gz emd_41616_validation.cif.gz | 20.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41616 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41616 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41616 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41616 | HTTPS FTP |
-Related structure data
| Related structure data |  8tu2MC  8tu0C  8tu1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41616.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41616.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.921 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41616_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
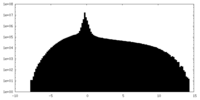
| Projections & Slices |
| ||||||||||||
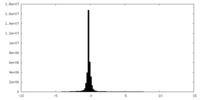
| Density Histograms |
-Half map: #2
| File | emd_41616_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
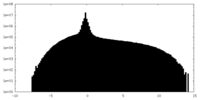
| Projections & Slices |
| ||||||||||||
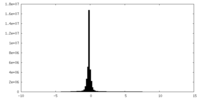
| Density Histograms |
- Sample components
Sample components
-Entire : Rat bocavirus
| Entire | Name:  Rat bocavirus Rat bocavirus |
|---|---|
| Components |
|
-Supramolecule #1: Rat bocavirus
| Supramolecule | Name: Rat bocavirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1788315 / Sci species name: Rat bocavirus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rat bocavirus Rat bocavirus |
| Molecular weight | Theoretical: 63.540734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MADGGESSNA NAGEGTSGGG GSGPLALTGG SRAGGTIGGG GAGSVGFSTG GWTAGTYFSD NKVVTNQTRQ FLTPIYNGHT YKALQADDL TNENDKSKWN GITTPWGYFN FNCYASHFSP QDWQRMCNEY KRWRPKRLRV QIYNLQLKTI QSNGADTQYN N DLTAAVHI ...String: MADGGESSNA NAGEGTSGGG GSGPLALTGG SRAGGTIGGG GAGSVGFSTG GWTAGTYFSD NKVVTNQTRQ FLTPIYNGHT YKALQADDL TNENDKSKWN GITTPWGYFN FNCYASHFSP QDWQRMCNEY KRWRPKRLRV QIYNLQLKTI QSNGADTQYN N DLTAAVHI LVDGSHQFPW AQHPWDDTCA PELPYVIYKT PQYAYFQNLA GLANNVGTNS ANKFLKMNTP LYVLETMSHE VL RTGEDTS FEFEMSSGWV DNQTNFCPPQ LDFNPLHDTR RVAPRATNNT TQYAPYPKFK KPSNWVPGPG MAYPGRGEAD GKR PAPMTV TLRPNTFIDA GNNTTDRFQQ ASYQEWKPTD DTIIGQSINV GPINCAATDP DAVTTAADAE DDVANPNTDK VSSH RYSID MTRWNAIQIN VRRNNGTPET TQIYRHYLYP MQAWNSNQID RYTPIWDKVP NTEWHTMLAS SDGTLPMTHP PGTIF IKCS KIPVPSENNA DSYLNIYCTG QVSYEIEWEC ERYNTKNWRP ELRVDPKNWT DPNNYNLNTQ GGYIVNEELY ETMPTK IGI NRVN UniProtKB: VP2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.52 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 9199 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)