+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CCT G beta 5 complex closed state 10 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CCT / Gb5 / complex / open / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationGTPase activator complex / light adaption / dark adaptation / G-protein gamma-subunit binding / negative regulation of voltage-gated calcium channel activity / zona pellucida receptor complex / scaRNA localization to Cajal body / chaperone mediated protein folding independent of cofactor / positive regulation of establishment of protein localization to telomere / positive regulation of protein localization to Cajal body ...GTPase activator complex / light adaption / dark adaptation / G-protein gamma-subunit binding / negative regulation of voltage-gated calcium channel activity / zona pellucida receptor complex / scaRNA localization to Cajal body / chaperone mediated protein folding independent of cofactor / positive regulation of establishment of protein localization to telomere / positive regulation of protein localization to Cajal body / tubulin complex assembly / chaperonin-containing T-complex / BBSome-mediated cargo-targeting to cilium / binding of sperm to zona pellucida / positive regulation of telomerase RNA localization to Cajal body / Folding of actin by CCT/TriC / Formation of tubulin folding intermediates by CCT/TriC / Prefoldin mediated transfer of substrate to CCT/TriC / RHOBTB1 GTPase cycle / intermediate filament cytoskeleton / WD40-repeat domain binding / G protein-coupled dopamine receptor signaling pathway / pericentriolar material / beta-tubulin binding / : / Association of TriC/CCT with target proteins during biosynthesis / chaperone-mediated protein complex assembly / RHOBTB2 GTPase cycle / heterochromatin / chaperone-mediated protein folding / protein folding chaperone / positive regulation of telomere maintenance via telomerase / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / GTPase activator activity / positive regulation of GTPase activity / acrosomal vesicle / cell projection / mRNA 3'-UTR binding / ATP-dependent protein folding chaperone / response to virus / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / cilium / G-protein activation / mRNA 5'-UTR binding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / azurophil granule lumen / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / unfolded protein binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / melanosome / protein folding / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / presynapse / protein-folding chaperone binding / Ca2+ pathway / cell body / G alpha (i) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / secretory granule lumen / ficolin-1-rich granule lumen / microtubule / Extra-nuclear estrogen signaling / cytoskeleton / protein stabilization / cadherin binding / G protein-coupled receptor signaling pathway / GTPase activity / centrosome / ubiquitin protein ligase binding / Neutrophil degranulation / Golgi apparatus / signal transduction / ATP hydrolysis activity / RNA binding / extracellular exosome / extracellular region / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
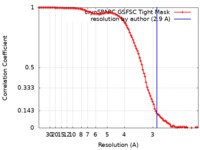
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Wang S / Sass M / Willardson BM / Shen PS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Visualizing the chaperone-mediated folding trajectory of the G protein β5 β-propeller. Authors: Shuxin Wang / Mikaila I Sass / Yujin Kwon / W Grant Ludlam / Theresa M Smith / Ethan J Carter / Nathan E Gladden / Margot Riggi / Janet H Iwasa / Barry M Willardson / Peter S Shen /  Abstract: The Chaperonin Containing Tailless polypeptide 1 (CCT) complex is an essential protein folding machine with a diverse clientele of substrates, including many proteins with β-propeller domains. Here, ...The Chaperonin Containing Tailless polypeptide 1 (CCT) complex is an essential protein folding machine with a diverse clientele of substrates, including many proteins with β-propeller domains. Here, we determine the structures of human CCT in complex with its accessory co-chaperone, phosducin-like protein 1 (PhLP1), in the process of folding Gβ, a component of Regulator of G protein Signaling (RGS) complexes. Cryoelectron microscopy (cryo-EM) and image processing reveal an ensemble of distinct snapshots that represent the folding trajectory of Gβ from an unfolded molten globule to a fully folded β-propeller. These structures reveal the mechanism by which CCT directs Gβ folding through initiating specific intermolecular contacts that facilitate the sequential folding of individual β sheets until the propeller closes into its native structure. This work directly visualizes chaperone-mediated protein folding and establishes that CCT orchestrates folding by stabilizing intermediates through interactions with surface residues that permit the hydrophobic core to coalesce into its folded state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40484.map.gz emd_40484.map.gz | 52.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40484-v30.xml emd-40484-v30.xml emd-40484.xml emd-40484.xml | 28.9 KB 28.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40484_fsc.xml emd_40484_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40484.png emd_40484.png | 578.2 KB | ||
| Filedesc metadata |  emd-40484.cif.gz emd-40484.cif.gz | 8.9 KB | ||
| Others |  emd_40484_half_map_1.map.gz emd_40484_half_map_1.map.gz emd_40484_half_map_2.map.gz emd_40484_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40484 http://ftp.pdbj.org/pub/emdb/structures/EMD-40484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40484 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40484 | HTTPS FTP |
-Validation report
| Summary document |  emd_40484_validation.pdf.gz emd_40484_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40484_full_validation.pdf.gz emd_40484_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_40484_validation.xml.gz emd_40484_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_40484_validation.cif.gz emd_40484_validation.cif.gz | 23.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40484 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40484 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40484 | HTTPS FTP |
-Related structure data
| Related structure data |  8shdMC  8sfeC  8sffC  8sg8C  8sg9C  8sgcC  8sglC  8sgqC  8sh9C  8shaC  8sheC  8shfC  8shgC  8shlC  8shnC  8shoC  8shpC  8shqC  8shtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40484.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40484.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||
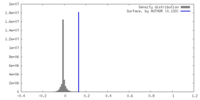
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_40484_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
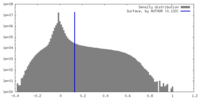
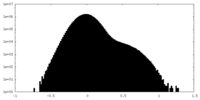
| Density Histograms |
-Half map: #2
| File | emd_40484_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
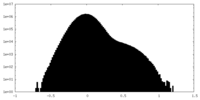
| Density Histograms |
- Sample components
Sample components
+Entire : CCT-Gb5-PhLP1 in closed state 10
+Supramolecule #1: CCT-Gb5-PhLP1 in closed state 10
+Macromolecule #1: Guanine nucleotide-binding protein subunit beta-5
+Macromolecule #2: T-complex protein 1 subunit alpha
+Macromolecule #3: T-complex protein 1 subunit beta
+Macromolecule #4: T-complex protein 1 subunit delta
+Macromolecule #5: T-complex protein 1 subunit epsilon
+Macromolecule #6: T-complex protein 1 subunit gamma
+Macromolecule #7: T-complex protein 1 subunit eta, N-terminally processed
+Macromolecule #8: T-complex protein 1 subunit theta
+Macromolecule #9: T-complex protein 1 subunit zeta
+Macromolecule #10: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #11: MAGNESIUM ION
+Macromolecule #12: ALUMINUM FLUORIDE
+Macromolecule #13: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK I | |||||||||||||||||||||
| Details | The sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.42 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)