+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CMKLR1 signaling complex | |||||||||
 Map data Map data | CMKLR1 complex map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Peptide agonist / Chemerin9 / Membrane protein / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationadipokinetic hormone binding / adipokinetic hormone receptor activity / complement receptor activity / chemokine receptor activity / Class A/1 (Rhodopsin-like receptors) / complement receptor mediated signaling pathway / negative regulation of interleukin-12 production / negative regulation of NF-kappaB transcription factor activity / positive regulation of macrophage chemotaxis / T cell migration ...adipokinetic hormone binding / adipokinetic hormone receptor activity / complement receptor activity / chemokine receptor activity / Class A/1 (Rhodopsin-like receptors) / complement receptor mediated signaling pathway / negative regulation of interleukin-12 production / negative regulation of NF-kappaB transcription factor activity / positive regulation of macrophage chemotaxis / T cell migration / positive regulation of fat cell differentiation / D2 dopamine receptor binding / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / G protein-coupled serotonin receptor binding / cellular response to forskolin / regulation of calcium-mediated signaling / regulation of mitotic spindle organization / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / skeletal system development / Regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / response to peptide hormone / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / chemotaxis / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / GDP binding / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / positive regulation of cold-induced thermogenesis / retina development in camera-type eye / signaling receptor activity / Ca2+ pathway / cell cortex / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / inflammatory response / immune response / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / centrosome / synapse / protein-containing complex binding / GTP binding / nucleolus / magnesium ion binding / signal transduction / extracellular exosome / nucleoplasm / membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Zhang X / Zhang C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Structural basis of G protein-Coupled receptor CMKLR1 activation and signaling induced by a chemerin-derived agonist. Authors: Xuan Zhang / Tina Weiß / Mary Hongying Cheng / Siqi Chen / Carla Katharina Ambrosius / Anne Sophie Czerniak / Kunpeng Li / Mingye Feng / Ivet Bahar / Annette G Beck-Sickinger / Cheng Zhang /   Abstract: Chemokine-like receptor 1 (CMKLR1), also known as chemerin receptor 23 (ChemR23) or chemerin receptor 1, is a chemoattractant G protein-coupled receptor (GPCR) that responds to the adipokine chemerin ...Chemokine-like receptor 1 (CMKLR1), also known as chemerin receptor 23 (ChemR23) or chemerin receptor 1, is a chemoattractant G protein-coupled receptor (GPCR) that responds to the adipokine chemerin and is highly expressed in innate immune cells, including macrophages and neutrophils. The signaling pathways of CMKLR1 can lead to both pro- and anti-inflammatory effects depending on the ligands and physiological contexts. To understand the molecular mechanisms of CMKLR1 signaling, we determined a high-resolution cryo-electron microscopy (cryo-EM) structure of the CMKLR1-Gi signaling complex with chemerin9, a nanopeptide agonist derived from chemerin, which induced complex phenotypic changes of macrophages in our assays. The cryo-EM structure, together with molecular dynamics simulations and mutagenesis studies, revealed the molecular basis of CMKLR1 signaling by elucidating the interactions at the ligand-binding pocket and the agonist-induced conformational changes. Our results are expected to facilitate the development of small molecule CMKLR1 agonists that mimic the action of chemerin9 to promote the resolution of inflammation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40450.map.gz emd_40450.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40450-v30.xml emd-40450-v30.xml emd-40450.xml emd-40450.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40450.png emd_40450.png | 75.4 KB | ||
| Filedesc metadata |  emd-40450.cif.gz emd-40450.cif.gz | 6.6 KB | ||
| Others |  emd_40450_half_map_1.map.gz emd_40450_half_map_1.map.gz emd_40450_half_map_2.map.gz emd_40450_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40450 http://ftp.pdbj.org/pub/emdb/structures/EMD-40450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40450 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40450 | HTTPS FTP |
-Validation report
| Summary document |  emd_40450_validation.pdf.gz emd_40450_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40450_full_validation.pdf.gz emd_40450_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_40450_validation.xml.gz emd_40450_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_40450_validation.cif.gz emd_40450_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40450 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40450 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40450 | HTTPS FTP |
-Related structure data
| Related structure data |  8sg1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40450.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40450.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CMKLR1 complex map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.848 Å | ||||||||||||||||||||||||||||||||||||
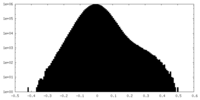
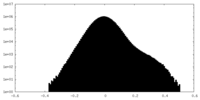
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: CMKLR1 complex map half A
| File | emd_40450_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CMKLR1 complex map half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
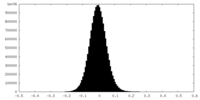
| Density Histograms |
-Half map: CMKLR1 complex map half B
| File | emd_40450_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CMKLR1 complex map half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
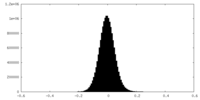
| Density Histograms |
- Sample components
Sample components
-Entire : Chemerin9-CMKLR1-Gi complex
| Entire | Name: Chemerin9-CMKLR1-Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: Chemerin9-CMKLR1-Gi complex
| Supramolecule | Name: Chemerin9-CMKLR1-Gi complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chemerin-like receptor 1
| Macromolecule | Name: Chemerin-like receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.079715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARVTRIFLVV VYSIVCFLGI LGNGLVIIIA TFKMKKTVNM VWFLNLAVAD FLFNVFLPIH ITYAAMDYHW VFGTAMCKIS NFLLIHNMF TSVFLLTIIS SDRCISVLLP VWSQNHRSVR LAYMACMVIW VLAFFLSSPS LVFRDTANLH GKISCFNNFS L STPGSSSW ...String: ARVTRIFLVV VYSIVCFLGI LGNGLVIIIA TFKMKKTVNM VWFLNLAVAD FLFNVFLPIH ITYAAMDYHW VFGTAMCKIS NFLLIHNMF TSVFLLTIIS SDRCISVLLP VWSQNHRSVR LAYMACMVIW VLAFFLSSPS LVFRDTANLH GKISCFNNFS L STPGSSSW PTHSQMDPVG YSRHMVVTVT RFLCGFLVPV LIITACYLTI VCKLQRNRLA KTKKPFKIIV TIIITFFLCW CP YHTLNLL ELHHTAMPGS VFSLGLPLAT ALAIANSCMN PILYVFMGQD FKKFK UniProtKB: Chemerin-like receptor 1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.956383 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWDS YTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD DNQIVTSSGD T TCALWDIE ...String: DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWDS YTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD DNQIVTSSGD T TCALWDIE TGQQTTTFTG HTGDVMSLSL APDTRLFVSG ACDASAKLWD VREGMCRQTF TGHESDINAI CFFPNGNAFA TG SDDATCR LFDLRADQEL MTYSHDNIIC GITSVSFSKS GRLLLAGYDD FNCNVWDALK ADRAGVLAGH DNRVSCLGVT DDG MAVATG SWDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: IAQARKLVQQ LKMEANIDRI KVSKAAADLM AYCEAHAKED PLLTPVPASQ NPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.12266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TLSAEDKAAV ERSKMIDRNL REDGEKAARE VKLLLLGAGE SGKNTIVKQM KIIHEAGYSE EECKQYKAVV YSNTIQSIIA IIRAMGRLK IDFGDSARAD DARQLFVLAG AAEEGFMTAE LAGVIKRLWK DSGVQACFNR SREYQLNDSA AYYLNDLDRI A QPNYIPTQ ...String: TLSAEDKAAV ERSKMIDRNL REDGEKAARE VKLLLLGAGE SGKNTIVKQM KIIHEAGYSE EECKQYKAVV YSNTIQSIIA IIRAMGRLK IDFGDSARAD DARQLFVLAG AAEEGFMTAE LAGVIKRLWK DSGVQACFNR SREYQLNDSA AYYLNDLDRI A QPNYIPTQ QDVLRTRVKT TGIVETHFTF KDLHFKMFDV GAQRSERKKW IHCFEGVTAI IFCVALSDYD LVLAEDEEMN RM HASMKLF DSICNNKWFT DTSIILFLNK KDLFEEKIKK SPLTICYPEY AGSNTYEEAA AYIQCQFEDL NKRKDTKEIY THF TCSTDT KNVQFVFDAV TDVIIKNNLK DCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #5: Chemerin 9
| Macromolecule | Name: Chemerin 9 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.063161 KDa |
| Sequence | String: YFPGQFAFS |
-Macromolecule #6: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.634797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL LEENLYFQGA SHHHHHHHH |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #8: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 8 / Number of copies: 2 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.94 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 242745 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)