[English] 日本語
 Yorodumi
Yorodumi- EMDB-40407: Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thio... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ligase / Signaling Protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationISG15 activating enzyme activity / Ligases; Forming carbon-sulfur bonds; Acid-thiol ligases / ISG15 transferase activity / positive regulation of protein oligomerization / ISG15-protein conjugation / regulation of type II interferon production / NS1 Mediated Effects on Host Pathways / protein localization to mitochondrion / response to type I interferon / Modulation of host responses by IFN-stimulated genes ...ISG15 activating enzyme activity / Ligases; Forming carbon-sulfur bonds; Acid-thiol ligases / ISG15 transferase activity / positive regulation of protein oligomerization / ISG15-protein conjugation / regulation of type II interferon production / NS1 Mediated Effects on Host Pathways / protein localization to mitochondrion / response to type I interferon / Modulation of host responses by IFN-stimulated genes / negative regulation of type I interferon-mediated signaling pathway / E2 ubiquitin-conjugating enzyme / negative regulation of viral genome replication / RSV-host interactions / ubiquitin conjugating enzyme activity / positive regulation of interleukin-10 production / positive regulation of bone mineralization / negative regulation of protein ubiquitination / positive regulation of interferon-beta production / positive regulation of erythrocyte differentiation / ubiquitin binding / integrin-mediated signaling pathway / Negative regulators of DDX58/IFIH1 signaling / protein modification process / Termination of translesion DNA synthesis / PKR-mediated signaling / DDX58/IFIH1-mediated induction of interferon-alpha/beta / modification-dependent protein catabolic process / integrin binding / ISG15 antiviral mechanism / response to virus / positive regulation of type II interferon production / protein tag activity / protein polyubiquitination / Interferon alpha/beta signaling / ubiquitin-protein transferase activity / Antigen processing: Ubiquitination & Proteasome degradation / defense response to virus / ubiquitin-dependent protein catabolic process / defense response to bacterium / protein ubiquitination / Amyloid fiber formation / innate immune response / DNA damage response / ubiquitin protein ligase binding / SARS-CoV-2 activates/modulates innate and adaptive immune responses / extracellular region / nucleoplasm / ATP binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
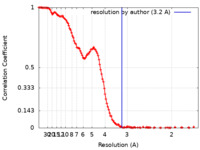
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Afsar M / Jia L / Ruben EA / Olsen SK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer. Authors: Mohammad Afsar / GuanQun Liu / Lijia Jia / Eliza A Ruben / Digant Nayak / Zuberwasim Sayyad / Priscila Dos Santos Bury / Kristin E Cano / Anindita Nayak / Xiang Ru Zhao / Ankita Shukla / ...Authors: Mohammad Afsar / GuanQun Liu / Lijia Jia / Eliza A Ruben / Digant Nayak / Zuberwasim Sayyad / Priscila Dos Santos Bury / Kristin E Cano / Anindita Nayak / Xiang Ru Zhao / Ankita Shukla / Patrick Sung / Elizabeth V Wasmuth / Michaela U Gack / Shaun K Olsen /  Abstract: ISG15 plays a crucial role in the innate immune response and has been well-studied due to its antiviral activity and regulation of signal transduction, apoptosis, and autophagy. ISG15 is a ubiquitin- ...ISG15 plays a crucial role in the innate immune response and has been well-studied due to its antiviral activity and regulation of signal transduction, apoptosis, and autophagy. ISG15 is a ubiquitin-like protein that is activated by an E1 enzyme (Uba7) and transferred to a cognate E2 enzyme (UBE2L6) to form a UBE2L6-ISG15 intermediate that functions with E3 ligases that catalyze conjugation of ISG15 to target proteins. Despite its biological importance, the molecular basis by which Uba7 catalyzes ISG15 activation and transfer to UBE2L6 is unknown as there is no available structure of Uba7. Here, we present cryo-EM structures of human Uba7 in complex with UBE2L6, ISG15 adenylate, and ISG15 thioester intermediate that are poised for catalysis of Uba7-UBE2L6-ISG15 thioester transfer. Our structures reveal a unique overall architecture of the complex compared to structures from the ubiquitin conjugation pathway, particularly with respect to the location of ISG15 thioester intermediate. Our structures also illuminate the molecular basis for Uba7 activities and for its exquisite specificity for ISG15 and UBE2L6. Altogether, our structural, biochemical, and human cell-based data provide significant insights into the functions of Uba7, UBE2L6, and ISG15 in cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40407.map.gz emd_40407.map.gz | 166.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40407-v30.xml emd-40407-v30.xml emd-40407.xml emd-40407.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40407_fsc.xml emd_40407_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_40407.png emd_40407.png | 21.7 KB | ||
| Masks |  emd_40407_msk_1.map emd_40407_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40407.cif.gz emd-40407.cif.gz | 7.1 KB | ||
| Others |  emd_40407_half_map_1.map.gz emd_40407_half_map_1.map.gz emd_40407_half_map_2.map.gz emd_40407_half_map_2.map.gz | 163.2 MB 163.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40407 http://ftp.pdbj.org/pub/emdb/structures/EMD-40407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40407 | HTTPS FTP |
-Related structure data
| Related structure data |  8se9MC  8seaC  8sebC  8sv8C  40410 C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40407.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40407.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8332 Å | ||||||||||||||||||||||||||||||||||||
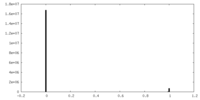
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40407_msk_1.map emd_40407_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
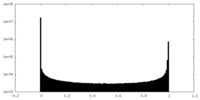
| Density Histograms |
-Half map: #1
| File | emd_40407_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
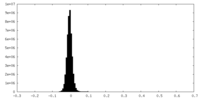
| Density Histograms |
-Half map: #2
| File | emd_40407_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
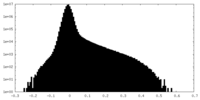
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thio...
| Entire | Name: Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thio...
| Supramolecule | Name: Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: Ubiquitin-like modifier-activating enzyme 7
| Macromolecule | Name: Ubiquitin-like modifier-activating enzyme 7 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 111.822102 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDALDASKLL DEELYSRQLY VLGSPAMQRI QGARVLVSGL QGLGAEVAKN LVLMGVGSLT LHDPHPTCWS DLAAQFLLSE QDLERSRAE ASQELLAQLN RAVQVVVHTG DITEDLLLDF QVVVLTAAKL EEQLKVGTLC HKHGVCFLAA DTRGLVGQLF C DFGEDFTV ...String: MDALDASKLL DEELYSRQLY VLGSPAMQRI QGARVLVSGL QGLGAEVAKN LVLMGVGSLT LHDPHPTCWS DLAAQFLLSE QDLERSRAE ASQELLAQLN RAVQVVVHTG DITEDLLLDF QVVVLTAAKL EEQLKVGTLC HKHGVCFLAA DTRGLVGQLF C DFGEDFTV QDPTEAEPLT AAIQHISQGS PGILTLRKGA NTHYFRDGDL VTFSGIEGMV ELNDCDPRSI HVREDGSLEI GD TTTFSRY LRGGAITEVK RPKTVRHKSL DTALLQPHVV AQSSQEVHHA HCLHQAFCAL HKFQHLHGRP PQPWDPVDAE TVV GLARDL EPLKRTEEEP LEEPLDEALV RTVALSSAGV LSPMVAMLGA VAAQEVLKAI SRKFMPLDQW LYFDALDCLP EDGE LLPSP EDCALRGSRY DGQIAVFGAG FQEKLRRQHY LLVGAGAIGC ELLKVFALVG LGAGNSGGLT VVDMDHIERS NLSRQ FLFR SQDVGRPKAE VAAAAARGLN PDLQVIPLTY PLDPTTEHIY GDNFFSRVDG VAAALDSFQA RRYVAARCTH YLKPLL EAG TSGTWGSATV FMPHVTEAYR APASAAASED APYPVCTVRY FPSTAEHTLQ WARHEFEELF RLSAETINHH QQAHTSL AD MDEPQTLTLL KPVLGVLRVR PQNWQDCVAW ALGHWKLCFH YGIKQLLRHF PPNKVLEDGT PFWSGPKQCP QPLEFDTN Q DTHLLYVLAA ANLYAQMHGL PGSQDWTALR ELLKLLPQPD PQQMAPIFAS NLELASASAE FGPEQQKELN KALEVWSVG PPLKPLMFEK DDDSNFHVDF VVAAASLRCQ NYGIPPVNRA QSKRIVGQII PAIATTTAAV AGLLGLELYK VVSGPRPRSA FRHSYLHLA ENYLIRYMPF APAIQTFHHL KWTSWDRLKV PAGQPERTLE SLLAHLQEQH GLRVRILLHG SALLYAAGWS P EKQAQHLP LRVTELVQQL TGQAPAPGQR VLVLELSCEG DDEDTAFPPL HYEL UniProtKB: Ubiquitin-like modifier-activating enzyme 7 |
-Macromolecule #2: Ubiquitin-like protein ISG15
| Macromolecule | Name: Ubiquitin-like protein ISG15 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.163701 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGWDLTVKML AGNEFQVSLS SSMSVSELKA QITQKIGVHA FQQRLAVHPS GVALQDRVPL ASQGLGPGST VLLVVDKCDE PLSILVRNN KGRSSTYEVR LTQTVAHLKQ QVSGLEGVQD DLFWLTFEGK PLEDQLPLGE YGLKPLSTVF MNLRLRGG UniProtKB: Ubiquitin-like protein ISG15 |
-Macromolecule #3: Ubiquitin/ISG15-conjugating enzyme E2 L6
| Macromolecule | Name: Ubiquitin/ISG15-conjugating enzyme E2 L6 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: E2 ubiquitin-conjugating enzyme |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.643377 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASMRVVKEL EDLQKKPPPY LRNLSSDDAN VLVWHALLLP DQPPYHLKAF NLRISFPPEY PFKPPMIKFT TKIYHPNVDE NGQICLPII SSENWKPSTK TSQVLEALNV LVNRPNIREP KRMDLADLLT QNPELFRKNA EEFTLRFGVD RPS UniProtKB: Ubiquitin/ISG15-conjugating enzyme E2 L6 |
-Macromolecule #4: ADENOSINE MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE MONOPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: AMP |
|---|---|
| Molecular weight | Theoretical: 347.221 Da |
| Chemical component information |  ChemComp-AMP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.888 µm / Nominal defocus min: 0.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)