+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Delta Spike in complex with JM-1A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Antibody / VIRAL PROTEIN-IMMUNE SYSTEM COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / membrane fusion / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
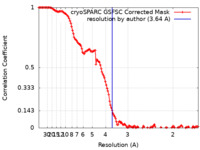
| Method | single particle reconstruction / cryo EM / Resolution: 3.64 Å | |||||||||
 Authors Authors | Nguyen VHT / Chen X | |||||||||
| Funding support |  Taiwan, 2 items Taiwan, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine. Authors: Xiaorui Chen / Arpita Mohapatra / Hong Thuy Vy Nguyen / Lisa Schimanski / Tiong Kit Tan / Pramila Rijal / Cheng-Pin Chen / Shu-Hsing Cheng / Wen-Hsin Lee / Yu-Chi Chou / Alain R Townsend / ...Authors: Xiaorui Chen / Arpita Mohapatra / Hong Thuy Vy Nguyen / Lisa Schimanski / Tiong Kit Tan / Pramila Rijal / Cheng-Pin Chen / Shu-Hsing Cheng / Wen-Hsin Lee / Yu-Chi Chou / Alain R Townsend / Che Ma / Kuan-Ying A Huang /   Abstract: Antibody-mediated immunity plays a key role in protection against SARS-CoV-2. We characterized B-cell-derived anti-SARS-CoV-2 RBD antibody repertoires from vaccinated and infected individuals and ...Antibody-mediated immunity plays a key role in protection against SARS-CoV-2. We characterized B-cell-derived anti-SARS-CoV-2 RBD antibody repertoires from vaccinated and infected individuals and elucidate the mechanism of action of broadly neutralizing antibodies and dissect antibodies at the epitope level. The breadth and clonality of anti-RBD B cell response varies among individuals. The majority of neutralizing antibody clones lose or exhibit reduced activities against Beta, Delta, and Omicron variants. Nevertheless, a portion of anti-RBD antibody clones that develops after a primary series or booster dose of COVID-19 vaccination exhibit broad neutralization against emerging Omicron BA.2, BA.4, BA.5, BQ.1.1, XBB.1.5 and XBB.1.16 variants. These broadly neutralizing antibodies share genetic features including a conserved usage of the IGHV3-53 and 3-9 genes and recognize three clustered epitopes of the RBD, including epitopes that partially overlap the classically defined set identified early in the pandemic. The Fab-RBD crystal and Fab-Spike complex structures corroborate the epitope grouping of antibodies and reveal the detailed binding mode of broadly neutralizing antibodies. Structure-guided mutagenesis improves binding and neutralization potency of antibody with Omicron variants via a single amino-substitution. Together, these results provide an immunological basis for partial protection against severe COVID-19 by the ancestral strain-based vaccine and indicate guidance for next generation monoclonal antibody development and vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39547.map.gz emd_39547.map.gz | 110.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39547-v30.xml emd-39547-v30.xml emd-39547.xml emd-39547.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39547_fsc.xml emd_39547_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_39547.png emd_39547.png | 119.8 KB | ||
| Filedesc metadata |  emd-39547.cif.gz emd-39547.cif.gz | 7.1 KB | ||
| Others |  emd_39547_half_map_1.map.gz emd_39547_half_map_1.map.gz emd_39547_half_map_2.map.gz emd_39547_half_map_2.map.gz | 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39547 http://ftp.pdbj.org/pub/emdb/structures/EMD-39547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39547 | HTTPS FTP |
-Validation report
| Summary document |  emd_39547_validation.pdf.gz emd_39547_validation.pdf.gz | 978.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39547_full_validation.pdf.gz emd_39547_full_validation.pdf.gz | 978.5 KB | Display | |
| Data in XML |  emd_39547_validation.xml.gz emd_39547_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_39547_validation.cif.gz emd_39547_validation.cif.gz | 28.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39547 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39547 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39547 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39547 | HTTPS FTP |
-Related structure data
| Related structure data |  8yrpMC  8x0xC  8x0yC  8yroC  8yz5C  8yz6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39547.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39547.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
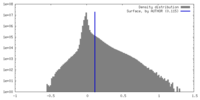
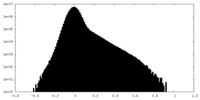
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39547_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
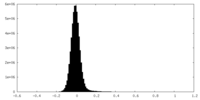
| Density Histograms |
-Half map: #1
| File | emd_39547_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
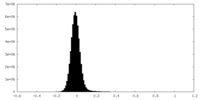
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Delta Spike in complex with JM-1A
| Entire | Name: SARS-CoV-2 Delta Spike in complex with JM-1A |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Delta Spike in complex with JM-1A
| Supramolecule | Name: SARS-CoV-2 Delta Spike in complex with JM-1A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 550 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 139.421547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVKLLVLLC TFTATYAGTQ CVNLRTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPV LPFNDGVYFA STEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV YYHKNNKSWM E SGVYSSAN ...String: MKVKLLVLLC TFTATYAGTQ CVNLRTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPV LPFNDGVYFA STEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV YYHKNNKSWM E SGVYSSAN NCTFEYVSQP FLMDLEGKQG NFKNLREFVF KNIDGYFKIY SKHTPINLVR DLPQGFSALE PLVDLPIGIN IT RFQTLLA LHRSYLTPGD SSSGWTAGAA AYYVGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTS NFRVQP TESIVRFPNI TNLCPFGEVF NATRFASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVY ADSFV IRGDEVRQIA PGQTGKIADY NYKLPDDFTG CVIAWNSNNL DSKVGGNYNY RYRLFRKSNL KPFERDISTE IYQAG SKPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESN KKF LPFQQFGRDI ADTTDAVRDP QTLEILDITP CSFGGVSVIT PGTNTSNQVA VLYQGVNCTE VPVAIHADQL TPTWRVY ST GSNVFQTRAG CLIGAEHVNN SYECDIPIGA GICASYQTQT NSRGSAGSVA SQSIIAYTMS LGAENSVAYS NNSIAIPT N FTISVTTEIL PVSMTKTSVD CTMYICGDST ECSNLLLQYG SFCTQLNRAL TGIAVEQDKN TQEVFAQVKQ IYKTPPIKD FGGFNFSQIL PDPSKPSKRS FIEDLLFNKV TLADAGFIKQ YGDCLGDIAA RDLICAQKFN GLTVLPPLLT DEMIAQYTSA LLAGTITSG WTFGAGAALQ IPFAMQMAYR FNGIGVTQNV LYENQKLIAN QFNSAIGKIQ DSLSSTASAL GKLQNVVNQN A QALNTLVK QLSSNFGAIS SVLNDILSRL DPPEAEVQID RLITGRLQSL QTYVTQQLIR AAEIRASANL AATKMSECVL GQ SKRVDFC GKGYHLMSFP QSAPHGVVFL HVTYVPAQEK NFTTAPAICH DGKAHFPREG VFVSNGTHWF VTQRNFYEPQ IIT TDNTFV SGNCDVVIGI VNNTVYDPLQ PELDSFKEEL DKYFKNHTSP DVDLGDISGI NASVVNIQKE IDRLNEVAKN LNES LIDLQ ELGKYEQDIR SLVPRGSPGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGHH HHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: JM-1A Heavy Chain
| Macromolecule | Name: JM-1A Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.411896 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQQSGPG LVKPSGTLSL TCAVSGASIS SGDWWSWVRQ SPGRGLEWIG GIFHSGTTNY SPSLKSRVTM SVDQPKNQFS LHLTSVTAA DTAVYYCARM RGIFDYWGQG TLVTVS |
-Macromolecule #3: JM-1A Light Chain
| Macromolecule | Name: JM-1A Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.27549 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQLTQSPSS LSVSVGDRVT ITCRASQAIS NSLAWYQQKP GKAPKLLLYA ASTLESGVPS RFSGSGSGTD FTLTISSLQP EDFATYYCQ HYYSTPFFGG GTKVEI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 258 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4422 / Average exposure time: 1.6 sec. / Average electron dose: 50.793 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)