+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Type I-EHNH Cascade complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA pro / RNA BINDING PROTEIN/RNA / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / hydrolase activity / RNA binding / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) | |||||||||
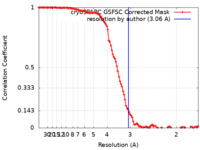
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Li Z | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems. Authors: Chendi Zhang / Fugen Chen / Feng Wang / Haijiang Xu / Jialin Xue / Zhuang Li /  Abstract: The metagenome-derived type I-E and type I-F variant CRISPR-associated complex for antiviral defense (Cascade) complexes, fused with HNH domains, precisely cleave target DNA, representing recently ...The metagenome-derived type I-E and type I-F variant CRISPR-associated complex for antiviral defense (Cascade) complexes, fused with HNH domains, precisely cleave target DNA, representing recently identified genome editing tools. However, the underlying working mechanisms remain unknown. Here, structures of type I-F and I-E Cascade complexes at different states are reported. In type I-F Cascade, Cas8f loosely attaches to Cascade head and is adjacent to the 5' end of the target single-stranded DNA (ssDNA). Formation of the full R-loop drives the Cascade head to move outward, allowing Cas8f to detach and rotate ∼150° to accommodate target ssDNA for cleavage. In type I-E Cascade, Cas5e domain is adjacent to the 5' end of the target ssDNA. Full crRNA-target pairing drives the lift of the Cascade head, widening the substrate channel for target ssDNA entrance. Altogether, these analyses into both complexes revealed that crRNA-guided positioning of target DNA and target DNA-induced HNH unlocking are two key factors for their site-specific cleavage of target DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39110.map.gz emd_39110.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39110-v30.xml emd-39110-v30.xml emd-39110.xml emd-39110.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39110_fsc.xml emd_39110_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_39110.png emd_39110.png | 105.4 KB | ||
| Filedesc metadata |  emd-39110.cif.gz emd-39110.cif.gz | 6.7 KB | ||
| Others |  emd_39110_half_map_1.map.gz emd_39110_half_map_1.map.gz emd_39110_half_map_2.map.gz emd_39110_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39110 http://ftp.pdbj.org/pub/emdb/structures/EMD-39110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39110 | HTTPS FTP |
-Validation report
| Summary document |  emd_39110_validation.pdf.gz emd_39110_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39110_full_validation.pdf.gz emd_39110_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_39110_validation.xml.gz emd_39110_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_39110_validation.cif.gz emd_39110_validation.cif.gz | 24.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39110 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39110 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39110 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39110 | HTTPS FTP |
-Related structure data
| Related structure data |  8yb6MC  8ydbC  8yeoC  8yh9C  8yhaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39110.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39110.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39110_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_39110_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Type I-EHNH Cascade complex
| Entire | Name: Type I-EHNH Cascade complex |
|---|---|
| Components |
|
-Supramolecule #1: Type I-EHNH Cascade complex
| Supramolecule | Name: Type I-EHNH Cascade complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) |
-Macromolecule #1: CRISPR system Cascade subunit CasD
| Macromolecule | Name: CRISPR system Cascade subunit CasD / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) |
| Molecular weight | Theoretical: 43.621383 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAPPNTLFL RLEGALQSWG SNEAKFALRR TADAPTKSGV LGLLCAAMGI GRAEAADSWL PKLANLRMGV RIDRPGIRWW DFHTVGAGQ RMRMAELKAP KKPSMVGAAL AETLTPSKVK TRAETLLSRR EYLADASFLV ALQGEPELVA KLSAALAKPV W AIYLGRKS ...String: MSAPPNTLFL RLEGALQSWG SNEAKFALRR TADAPTKSGV LGLLCAAMGI GRAEAADSWL PKLANLRMGV RIDRPGIRWW DFHTVGAGQ RMRMAELKAP KKPSMVGAAL AETLTPSKVK TRAETLLSRR EYLADASFLV ALQGEPELVA KLSAALAKPV W AIYLGRKS CPPSRPVCEH PPGFYNTLEE ALSAVPLQKR WHNEPLPQIL PCVMDWIPGY DGEHAPDDAE IHYDLPVSFQ PP RHLPRFV IRRELVVGED VQVSRETGTS VWRPKGTRAD YNNSEYKKVR AERLVMDHAA CMVCKAPATT VQHVNYRRAG GKE IPEDLR ALCRLCHDAC TMLEYGSGMT TNRIDPCDPI WRERILAKRK EIVEFRSRGQ RFRKMKPEEE NG UniProtKB: CRISPR system Cascade subunit CasD |
-Macromolecule #2: CRISPR-associated endoribonuclease Cse3
| Macromolecule | Name: CRISPR-associated endoribonuclease Cse3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) |
| Molecular weight | Theoretical: 31.218768 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIYLSRLLID TGGNPDRPRP GRKWLDNIYN VHRRLSMAFP SGLRREQDPH FLKPFSPNDF QKTPFLFRVD NNIDGNDKRA IIIVQSVLE PDWDYCFQNA LDFLAAPPET KEYNPEFKAG QLLRFRLRVN ASVRRHIPEM VQQDGQTIET GKILHKRVSL T WDASSTPD ...String: MIYLSRLLID TGGNPDRPRP GRKWLDNIYN VHRRLSMAFP SGLRREQDPH FLKPFSPNDF QKTPFLFRVD NNIDGNDKRA IIIVQSVLE PDWDYCFQNA LDFLAAPPET KEYNPEFKAG QLLRFRLRVN ASVRRHIPEM VQQDGQTIET GKILHKRVSL T WDASSTPD QALADWLAAK SPKLGFTLQR CELLQLGWVY GSKPEPKNVK VKEQGQGYWR EHKYNPLRFR AALLEGVLEV DD PKLFLKT LSSGIGKAKS FGFGLLSVLP IRNDG UniProtKB: CRISPR-associated endoribonuclease Cse3 |
-Macromolecule #4: CRISPR system Cascade subunit CasC
| Macromolecule | Name: CRISPR system Cascade subunit CasC / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) |
| Molecular weight | Theoretical: 41.794367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLIEIHMIQN HSPANLNRDD LGAPKTCYFG GVLRSRISSQ CIKRSIRTSN DFKALLGGVR TRRLADLIQQ EAGETECWKK AQEILNKCG FKNKDDNTKM LVFMSKDKIK DLARIVLDNS LGLTEAAQQV ANVIAQATLA PDIALCGRML EPNDKDKDKK V KWSNTTVE ...String: MLIEIHMIQN HSPANLNRDD LGAPKTCYFG GVLRSRISSQ CIKRSIRTSN DFKALLGGVR TRRLADLIQQ EAGETECWKK AQEILNKCG FKNKDDNTKM LVFMSKDKIK DLARIVLDNS LGLTEAAQQV ANVIAQATLA PDIALCGRML EPNDKDKDKK V KWSNTTVE AALQVAHAIS THIARPEIDY FVAADDVPGE DAGAGHIGES MFASACFYKY FSIDWEQLVK NLKGDTNLAA HT VGAFLLA AAKTNPSGKQ NSFAAHNYPD GILVEFKNSP ISYANAFVRP VSVVKESDLV EQSIGQLSNY VNDIRLGYYD EQS PVIGFW FSPNNRYPLG YKHSKLASRN IGNLNELVGA VLDYIGGFKW EEVQKSKAYI GG UniProtKB: CRISPR system Cascade subunit CasC |
-Macromolecule #5: CRISPR-associated protein Cse1 (CRISPR_cse1)
| Macromolecule | Name: CRISPR-associated protein Cse1 (CRISPR_cse1) / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) |
| Molecular weight | Theoretical: 60.665359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYCAAVCFPQ TKYQQRGTAL KKPLVGLRKM GVEAAAWNTL KVTRDRPKLT FPDLITPQSK FLDNDLWLKY KVPIEQKEHV MYNLLCDNW VNVVYLSGKP DRISLVQTLK DAHCLQLAYS NPMDRFTVFR FLLALGYWCF ANTNVEPEPD KPLPVSWIPW L EENKEYFE ...String: MYCAAVCFPQ TKYQQRGTAL KKPLVGLRKM GVEAAAWNTL KVTRDRPKLT FPDLITPQSK FLDNDLWLKY KVPIEQKEHV MYNLLCDNW VNVVYLSGKP DRISLVQTLK DAHCLQLAYS NPMDRFTVFR FLLALGYWCF ANTNVEPEPD KPLPVSWIPW L EENKEYFE LFGDGKRFFQ ADPSSRIRAI TDLIHEIPTA HNLCHFKHVT DYIDGLCEAC CIKGLLRLPV FTTVGGRGIG AG INNTPPF YLLWHANDLA GMLAQNWQPW DNMGIPAWLG SFQKESREVG LLAGMTWLPR KVYLHDPVPG QAACCSCGLP SEA LVYSCS IEVEPVPKGL EWKDPHGVYT DQGKSLQSKI KLMSNDRYTF ADRDWYSPLF SYLHAEGNSR QGKLWLVGFA SDKA KSIDI WDKIIELEGT DTNDELLAQL ANRATALNAM RKKPLRGDFK KSVGTPQIAD IIPHAENRIA INAGKMTENR GYSWQ DADT EYGELLTKVA YSLEPAQTVD ARLKRGNFIS RKPWPIIPES KTKPAEGDQN E UniProtKB: CRISPR-associated protein Cse1 (CRISPR_cse1) |
-Macromolecule #6: CRISPR-associated protein Cse2 (CRISPR_cse2)
| Macromolecule | Name: CRISPR-associated protein Cse2 (CRISPR_cse2) / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) |
| Molecular weight | Theoretical: 20.300639 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNRGTVDFIA SLENLKEGDL GILRKLRGAR LDEKLPGFDL FSALWWPLRQ KNQRAPKREV AWLIAKLFAE FRFEQREGAT LPILMGGIC RKLEPKKELP RVLARFDQLA SLDIMQMEEP LSVIMGILRK HQQVCLDWVG LTDVLSFWEQ EPVKREWSDS F IKAYKINK EDSDVD UniProtKB: CRISPR-associated protein Cse2 (CRISPR_cse2) |
-Macromolecule #3: 61-nt crRNA
| Macromolecule | Name: 61-nt crRNA / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) Candidatus Cloacimonetes bacterium ADurb.Bin088 (bacteria) |
| Molecular weight | Theoretical: 19.681744 KDa |
| Sequence | String: GUGAACCGGA GAAGUCAUUU AAUAAGGCCA CUGUUAAAAA GUAUUCCCCA CGCAUGUGGG G |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)