[English] 日本語
 Yorodumi
Yorodumi- EMDB-37098: Complex of DDM1-nucleosome(H2A) complex with DDM1 bound to SHL2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of DDM1-nucleosome(H2A) complex with DDM1 bound to SHL2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DDM1 / nucleosome / H2A / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-mediated transformation / retrotransposition / chromocenter / response to water deprivation / plasmodesma / plant-type vacuole / thylakoid / DNA methylation-dependent constitutive heterochromatin formation / ATP-dependent chromatin remodeler activity / plastid ...DNA-mediated transformation / retrotransposition / chromocenter / response to water deprivation / plasmodesma / plant-type vacuole / thylakoid / DNA methylation-dependent constitutive heterochromatin formation / ATP-dependent chromatin remodeler activity / plastid / DNA helicase activity / epigenetic regulation of gene expression / chloroplast / response to bacterium / heterochromatin formation / response to wounding / structural constituent of chromatin / nucleosome / peroxisome / DNA helicase / chromatin remodeling / protein heterodimerization activity / nucleolus / ATP hydrolysis activity / DNA binding / extracellular region / ATP binding / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
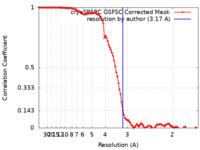
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Zhang H / Zhang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Mechanism of heterochromatin remodeling revealed by the DDM1 bound nucleosome structures. Authors: Hongwei Zhang / Zhanxi Gu / Yuan Zeng / Yu Zhang /  Abstract: The SWI/SNF2 chromatin remodeling factor decreased DNA methylation 1 (DDM1) is essential for the silencing of transposable elements (TEs) in both euchromatic and heterochromatic regions. Here, we ...The SWI/SNF2 chromatin remodeling factor decreased DNA methylation 1 (DDM1) is essential for the silencing of transposable elements (TEs) in both euchromatic and heterochromatic regions. Here, we determined the cryo-EM structures of DDM1-nucleosome and DDM1-nucleosome complexes at near-atomic resolution in the presence of the ATP analog ADP-BeFx. The structures show that nucleosomal DNA is unwrapped more on the surface of the histone octamer containing histone H2A than that containing histone H2A.W. DDM1 embraces one DNA gyre of the nucleosome and interacts with the N-terminal tails of histone H4. Although we did not observe DDM1-H2A.W interactions in our structures, the results of the pull-down experiments suggest a direct interaction between DDM1 and the core region of histone H2A.W. Our work provides mechanistic insights into the heterochromatin remodeling process driven by DDM1 in plants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37098.map.gz emd_37098.map.gz | 51.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37098-v30.xml emd-37098-v30.xml emd-37098.xml emd-37098.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37098_fsc.xml emd_37098_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_37098.png emd_37098.png | 41.8 KB | ||
| Filedesc metadata |  emd-37098.cif.gz emd-37098.cif.gz | 7.1 KB | ||
| Others |  emd_37098_half_map_1.map.gz emd_37098_half_map_1.map.gz emd_37098_half_map_2.map.gz emd_37098_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37098 http://ftp.pdbj.org/pub/emdb/structures/EMD-37098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37098 | HTTPS FTP |
-Validation report
| Summary document |  emd_37098_validation.pdf.gz emd_37098_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37098_full_validation.pdf.gz emd_37098_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_37098_validation.xml.gz emd_37098_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_37098_validation.cif.gz emd_37098_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37098 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37098 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37098 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37098 | HTTPS FTP |
-Related structure data
| Related structure data |  8kcbMC  8kccC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37098.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37098.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37098_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
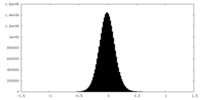
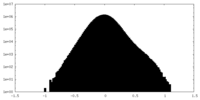
| Density Histograms |
-Half map: #1
| File | emd_37098_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
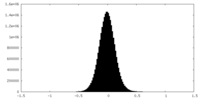
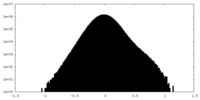
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of DDM1-nucleosome(H2A.W) complex with DDM1 bound to SHL2
+Supramolecule #1: Complex of DDM1-nucleosome(H2A.W) complex with DDM1 bound to SHL2
+Macromolecule #1: Histone H2B.10
+Macromolecule #2: Histone H3.1
+Macromolecule #3: Histone H4
+Macromolecule #4: Histone H2A.6
+Macromolecule #7: ATP-dependent DNA helicase DDM1
+Macromolecule #5: DNA (170-MER)
+Macromolecule #6: DNA (170-MER)
+Macromolecule #8: BERYLLIUM TRIFLUORIDE ION
+Macromolecule #9: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)