+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human ATAD2 Walker B mutant-H3/H4K5Q complex, ATP state | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Histone chaperone / AAA+ ATPase / GENE REGULATION | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnucleosome disassembly / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / transcription initiation-coupled chromatin remodeling / nucleosome assembly / histone binding / chromatin binding / positive regulation of DNA-templated transcription / ATP hydrolysis activity / extracellular exosome ...nucleosome disassembly / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / transcription initiation-coupled chromatin remodeling / nucleosome assembly / histone binding / chromatin binding / positive regulation of DNA-templated transcription / ATP hydrolysis activity / extracellular exosome / nucleoplasm / ATP binding / nucleus Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
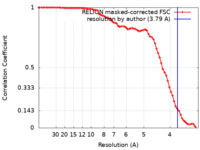
| Method | single particle reconstruction / cryo EM / Resolution: 3.79 Å | ||||||||||||||||||
 Authors Authors | Cho C / Song J | ||||||||||||||||||
| Funding support |  Korea, Republic Of, 5 items Korea, Republic Of, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication. Authors: Carol Cho / Christian Ganser / Takayuki Uchihashi / Koichi Kato / Ji-Joon Song /   Abstract: ATAD2 is a non-canonical ATP-dependent histone chaperone and a major cancer target. Despite widespread efforts to design drugs targeting the ATAD2 bromodomain, little is known about the overall ...ATAD2 is a non-canonical ATP-dependent histone chaperone and a major cancer target. Despite widespread efforts to design drugs targeting the ATAD2 bromodomain, little is known about the overall structural organization and regulation of ATAD2. Here, we present the 3.1 Å cryo-EM structure of human ATAD2 in the ATP state, showing a shallow hexameric spiral that binds a peptide substrate at the central pore. The spiral conformation is locked by an N-terminal linker domain (LD) that wedges between the seam subunits, thus limiting ATP-dependent symmetry breaking of the AAA+ ring. In contrast, structures of the ATAD2-histone H3/H4 complex show the LD undocked from the seam, suggesting that H3/H4 binding unlocks the AAA+ spiral by allosterically releasing the LD. These findings, together with the discovery of an inter-subunit signaling mechanism, reveal a unique regulatory mechanism for ATAD2 and lay the foundation for developing new ATAD2 inhibitors. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36665.map.gz emd_36665.map.gz | 8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36665-v30.xml emd-36665-v30.xml emd-36665.xml emd-36665.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36665_fsc.xml emd_36665_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_36665.png emd_36665.png | 34.7 KB | ||
| Filedesc metadata |  emd-36665.cif.gz emd-36665.cif.gz | 5.9 KB | ||
| Others |  emd_36665_half_map_1.map.gz emd_36665_half_map_1.map.gz emd_36665_half_map_2.map.gz emd_36665_half_map_2.map.gz | 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36665 http://ftp.pdbj.org/pub/emdb/structures/EMD-36665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36665 | HTTPS FTP |
-Validation report
| Summary document |  emd_36665_validation.pdf.gz emd_36665_validation.pdf.gz | 705.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36665_full_validation.pdf.gz emd_36665_full_validation.pdf.gz | 704.8 KB | Display | |
| Data in XML |  emd_36665_validation.xml.gz emd_36665_validation.xml.gz | 15.2 KB | Display | |
| Data in CIF |  emd_36665_validation.cif.gz emd_36665_validation.cif.gz | 20.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36665 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36665 | HTTPS FTP |
-Related structure data
| Related structure data |  8juwMC  8h3hC  8juyC  8juzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36665.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36665.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
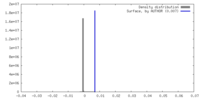
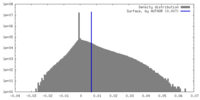
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36665_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
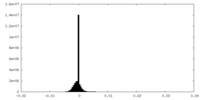
| Density Histograms |
-Half map: #2
| File | emd_36665_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
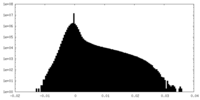
| Density Histograms |
- Sample components
Sample components
-Entire : ATAD2
| Entire | Name: ATAD2 |
|---|---|
| Components |
|
-Supramolecule #1: ATAD2
| Supramolecule | Name: ATAD2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATPase family AAA domain-containing protein 2
| Macromolecule | Name: ATPase family AAA domain-containing protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.690406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DRMKIGASLA DVDPMQLDSS VRFDSVGGLS NHIAALKEMV VFPLLYPEVF EKFKIQPPRG CLFYGPPGTG KTLVARALAN ECSQGDKRV AFFMRKGADC LSKWVGESER QLRLLFDQAY QMRPSIIFFD QIDGLAPVRS SRQDQIHSSI VSTLLALMDG L DSRGEIVV ...String: DRMKIGASLA DVDPMQLDSS VRFDSVGGLS NHIAALKEMV VFPLLYPEVF EKFKIQPPRG CLFYGPPGTG KTLVARALAN ECSQGDKRV AFFMRKGADC LSKWVGESER QLRLLFDQAY QMRPSIIFFD QIDGLAPVRS SRQDQIHSSI VSTLLALMDG L DSRGEIVV IGATNRLDSI DPALRRPGRF DREFLFSLPD KEARKEILKI HTRDWNPKPL DTFLEELAEN CVGYCGADIK SI CAEAALC ALRRRYPQIY TTSEKLQLDL SSINISAKDF EVAMQKMIPA SQRAVTSPGQ ALSTVVKPLL QNTVDKILEA LQR VFPHAE FRTNKTLDSD ISCPLLESDL AYSDDDVPSV YENGLSQKSS HKAKDNFNFL HLNRNACYQP MSFRPRILIV GEPG FGQGS HLAPAVIHAL EKFTVYTLDI PVLFGVSTTS PEETCAQVIR EAKRTAPSIV YVPHIHVWWE IVGPTLKATF TTLLQ NIPS FAPVLLLATS DKPHSALPEE VQELFIRDYG EIFNVQLPDK EERTKFFEDL ILKQAAKPPI SKKKAVLQAL EVLPVA PPP EPRSLTAEEV KRLEEQEEYA PSYYHVMPKQ NSTLVGDKRS DPEQNEKLKT PSTPVACSTP AQLKRKIRKK SNWYLGT IK KRRKISQAKD DSQNAIDHKI ESDTEETQDT SVDHNETGNT GESSVEENEK QQNASESKLE LRNNSNTCNI ENELEDSR K TTACTELRDK IACNGDASSS QIIHISDENE GKEMCVLRMT QPTPSLVVDH ERLKNLLKTV VKKSQNYNIF QLENLYAVI SQCIYRHRKD HDKTSLIQKM EQEVENFSCS R UniProtKB: ATPase family AAA domain-containing protein 2, ATPase family AAA domain-containing protein 2, ATPase family AAA domain-containing protein 2 |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)