[English] 日本語
 Yorodumi
Yorodumi- EMDB-34924: The cryo-EM structure of cellobiose phosphorylase from Clostridiu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of cellobiose phosphorylase from Clostridium thermocellum ( varient) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cellobiose phosphorylase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellobiose phosphorylase / cellobiose phosphorylase activity / cellulose catabolic process / carbohydrate binding Similarity search - Function | |||||||||
| Biological species |  Acetivibrio thermocellus (bacteria) Acetivibrio thermocellus (bacteria) | |||||||||
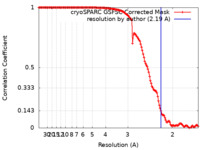
| Method | single particle reconstruction / cryo EM / Resolution: 2.19 Å | |||||||||
 Authors Authors | Iriya S / Kuga T / Sunagawa N / Igarashi K | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The cryo-EM structure of cellobiose phosphorylase from Clostridium thermocellum Authors: Iriya S / Kuga T / Sunagawa N / Igarashi K | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34924.map.gz emd_34924.map.gz | 219.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34924-v30.xml emd-34924-v30.xml emd-34924.xml emd-34924.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34924_fsc.xml emd_34924_fsc.xml | 13.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_34924.png emd_34924.png | 110.8 KB | ||
| Masks |  emd_34924_msk_1.map emd_34924_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34924.cif.gz emd-34924.cif.gz | 6.1 KB | ||
| Others |  emd_34924_half_map_1.map.gz emd_34924_half_map_1.map.gz emd_34924_half_map_2.map.gz emd_34924_half_map_2.map.gz | 226.7 MB 226.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34924 http://ftp.pdbj.org/pub/emdb/structures/EMD-34924 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34924 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34924 | HTTPS FTP |
-Validation report
| Summary document |  emd_34924_validation.pdf.gz emd_34924_validation.pdf.gz | 904.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34924_full_validation.pdf.gz emd_34924_full_validation.pdf.gz | 904.2 KB | Display | |
| Data in XML |  emd_34924_validation.xml.gz emd_34924_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_34924_validation.cif.gz emd_34924_validation.cif.gz | 27.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34924 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34924 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34924 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34924 | HTTPS FTP |
-Related structure data
| Related structure data |  8hobMC  8ho7C  8ho9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34924.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34924.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
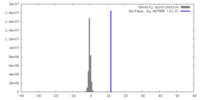
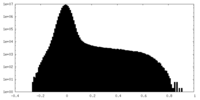
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34924_msk_1.map emd_34924_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
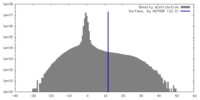
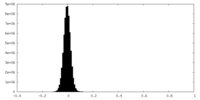
| Density Histograms |
-Half map: #1
| File | emd_34924_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
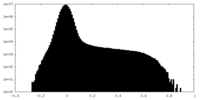
| Density Histograms |
-Half map: #2
| File | emd_34924_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
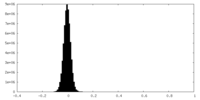
| Density Histograms |
- Sample components
Sample components
-Entire : Cellobiose phosphorylase
| Entire | Name: Cellobiose phosphorylase |
|---|---|
| Components |
|
-Supramolecule #1: Cellobiose phosphorylase
| Supramolecule | Name: Cellobiose phosphorylase / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Acetivibrio thermocellus (bacteria) Acetivibrio thermocellus (bacteria) |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Cellobiose phosphorylase
| Macromolecule | Name: Cellobiose phosphorylase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: cellobiose phosphorylase |
|---|---|
| Source (natural) | Organism:  Acetivibrio thermocellus (bacteria) Acetivibrio thermocellus (bacteria) |
| Molecular weight | Theoretical: 93.973258 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKFGFFDDA NKEYVITVPR TPYPWINYLG TENFFSLISN TAGGYSFYRD ARLRRITRYR YNNVPIDMGG RYFYIYDNGD FWSPGWSPV KRELESYESR HGLGYTKIAG KRNGIKAEVT FFVPLNYNGE VQKLILKNEG QDKKKITLFS FIEFSLWNAY D DMTNFQRN ...String: MGKFGFFDDA NKEYVITVPR TPYPWINYLG TENFFSLISN TAGGYSFYRD ARLRRITRYR YNNVPIDMGG RYFYIYDNGD FWSPGWSPV KRELESYESR HGLGYTKIAG KRNGIKAEVT FFVPLNYNGE VQKLILKNEG QDKKKITLFS FIEFSLWNAY D DMTNFQRN FSTGEVEIEG SVIYHKTEYR ERRNHYAFYS VNAKISGFDS DRDSFIGLYN GFDAPQAVVN GKSNNSVADG WA PIASHSI EIELNPGEQK EYVFIIGYVE NKDEEKWESK GVINKKKAYE MIEQFNTVEK VDKAFEELKS YWNALLSKYF LES HDEKLN RMVNIWNQYQ SMVTFNMSRS ASYFESGIGR GMGFRDSNQD LLGFVHQIPE RARERLLDLA ATQLEDGSAY HQYQ PLTKK GNNEIGSNFN DDPLWLILAT AAYIKETGDY SILKEQVPFN NDPSKADTMF EHLTRSFYHV VNNLGPHGLP LIGRA DWND CLNLNCFSTV PDESFQTTTS KDGKVAESVM IAGMFVFIGK DYVKLSEYMG LEEEARKAQQ HIDAMKEAIL KYGYDG EWF LRAYDDFGRK VGSKENEEGK IFIESQGFSV MAEIGLEDGK ALKALDSVKK YLDTPYGLVL QNPAFTRYYI EYGEIST YP PGYKENAGIF SHNNAWIISA ETVVGRGDMA FDYYRKIAPA YIEDVSDIHK LEPYVYAQMV AGKDAKRHGE AKNSWLTG T AAWNFVAISQ WILGVKPDYD GLKIDPSIPK AWDGYKVTRY FRGSTYEITV KNPNHVSKGV AKITVDGNEI SGNILPVFN DGKTHKVEVI MGLEHHHHHH UniProtKB: Cellobiose phosphorylase |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 333 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
Details: 20 mM MES, 70 mM NaCl | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV Details: Vitrification carried out in nitrogen atmosphere.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 3744 / Average exposure time: 5.567 sec. / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)