+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outer arm dynein of zebrafish sperm axoneme, WT | ||||||||||||
 Map data Map data | outer arm dynein, WT zebrafish sperm axoneme | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
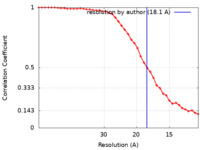
| Method | subtomogram averaging / cryo EM / Resolution: 18.1 Å | ||||||||||||
 Authors Authors | Yamaguchi H / Kikkawa M | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Calaxin stabilizes the docking of outer arm dyneins onto ciliary doublet microtubule in vertebrates. Authors: Hiroshi Yamaguchi / Motohiro Morikawa / Masahide Kikkawa /  Abstract: Outer arm dynein (OAD) is the main force generator of ciliary beating. Although OAD loss is the most frequent cause of human primary ciliary dyskinesia, the docking mechanism of OAD onto the ciliary ...Outer arm dynein (OAD) is the main force generator of ciliary beating. Although OAD loss is the most frequent cause of human primary ciliary dyskinesia, the docking mechanism of OAD onto the ciliary doublet microtubule (DMT) remains elusive in vertebrates. Here, we analyzed the functions of Calaxin/Efcab1 and Armc4, the two of five components of vertebrate OAD-DC (docking complex), using zebrafish spermatozoa and cryo-electron tomography. Mutation of caused complete loss of OAD, whereas mutation of caused only partial loss of OAD. Detailed structural analysis revealed that OADs are tethered to DMT through DC components other than Calaxin, and that recombinant Calaxin can autonomously rescue the deficient DC structure and the OAD instability. Our data demonstrate the discrete roles of Calaxin and Armc4 in the OAD-DMT interaction, suggesting the stabilizing process of OAD docking onto DMT in vertebrates. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34798.map.gz emd_34798.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34798-v30.xml emd-34798-v30.xml emd-34798.xml emd-34798.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34798_fsc.xml emd_34798_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_34798.png emd_34798.png | 61.9 KB | ||
| Masks |  emd_34798_msk_1.map emd_34798_msk_1.map | 3.8 MB |  Mask map Mask map | |
| Others |  emd_34798_additional_1.map.gz emd_34798_additional_1.map.gz emd_34798_half_map_1.map.gz emd_34798_half_map_1.map.gz emd_34798_half_map_2.map.gz emd_34798_half_map_2.map.gz | 1.2 MB 3.2 MB 3.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34798 http://ftp.pdbj.org/pub/emdb/structures/EMD-34798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34798 | HTTPS FTP |
-Validation report
| Summary document |  emd_34798_validation.pdf.gz emd_34798_validation.pdf.gz | 853.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34798_full_validation.pdf.gz emd_34798_full_validation.pdf.gz | 853.5 KB | Display | |
| Data in XML |  emd_34798_validation.xml.gz emd_34798_validation.xml.gz | 9.9 KB | Display | |
| Data in CIF |  emd_34798_validation.cif.gz emd_34798_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34798 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34798 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34798 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34798 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34798.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34798.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | outer arm dynein, WT zebrafish sperm axoneme | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.14 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34798_msk_1.map emd_34798_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: masked
| File | emd_34798_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | masked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map1, unmasked
| File | emd_34798_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map1, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map2, unmasked
| File | emd_34798_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map2, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Outer arm dynein of zebrafish sperm axoneme, WT
| Entire | Name: Outer arm dynein of zebrafish sperm axoneme, WT |
|---|---|
| Components |
|
-Supramolecule #1: Outer arm dynein of zebrafish sperm axoneme, WT
| Supramolecule | Name: Outer arm dynein of zebrafish sperm axoneme, WT / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Details: 30 mM HEPES at pH 7.2, 5 mM MgSO4, 1 mM dithiothreitol, 1 mM EGTA, and 50 mM CH3COOK |
|---|---|
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 35 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 2.45 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 15000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)