+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | rO44 A-form (staple xover asymmetry) | |||||||||
 Map data Map data | rO44 A-form (staple xover asymmetry) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Inovirus M13 Inovirus M13 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 17.0 Å | |||||||||
 Authors Authors | Zhang K / Li S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: 3D RNA-scaffolded wireframe origami. Authors: Molly F Parsons / Matthew F Allan / Shanshan Li / Tyson R Shepherd / Sakul Ratanalert / Kaiming Zhang / Krista M Pullen / Wah Chiu / Silvi Rouskin / Mark Bathe /   Abstract: Hybrid RNA:DNA origami, in which a long RNA scaffold strand folds into a target nanostructure via thermal annealing with complementary DNA oligos, has only been explored to a limited extent despite ...Hybrid RNA:DNA origami, in which a long RNA scaffold strand folds into a target nanostructure via thermal annealing with complementary DNA oligos, has only been explored to a limited extent despite its unique potential for biomedical delivery of mRNA, tertiary structure characterization of long RNAs, and fabrication of artificial ribozymes. Here, we investigate design principles of three-dimensional wireframe RNA-scaffolded origami rendered as polyhedra composed of dual-duplex edges. We computationally design, fabricate, and characterize tetrahedra folded from an EGFP-encoding messenger RNA and de Bruijn sequences, an octahedron folded with M13 transcript RNA, and an octahedron and pentagonal bipyramids folded with 23S ribosomal RNA, demonstrating the ability to make diverse polyhedral shapes with distinct structural and functional RNA scaffolds. We characterize secondary and tertiary structures using dimethyl sulfate mutational profiling and cryo-electron microscopy, revealing insight into both global and local, base-level structures of origami. Our top-down sequence design strategy enables the use of long RNAs as functional scaffolds for complex wireframe origami. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34058.map.gz emd_34058.map.gz | 26.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34058-v30.xml emd-34058-v30.xml emd-34058.xml emd-34058.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34058.png emd_34058.png | 45 KB | ||
| Others |  emd_34058_half_map_1.map.gz emd_34058_half_map_1.map.gz emd_34058_half_map_2.map.gz emd_34058_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34058 http://ftp.pdbj.org/pub/emdb/structures/EMD-34058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34058 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34058 | HTTPS FTP |
-Validation report
| Summary document |  emd_34058_validation.pdf.gz emd_34058_validation.pdf.gz | 682.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34058_full_validation.pdf.gz emd_34058_full_validation.pdf.gz | 682.4 KB | Display | |
| Data in XML |  emd_34058_validation.xml.gz emd_34058_validation.xml.gz | 11.1 KB | Display | |
| Data in CIF |  emd_34058_validation.cif.gz emd_34058_validation.cif.gz | 13 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34058 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34058 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34058 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34058 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34058.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34058.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rO44 A-form (staple xover asymmetry) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||
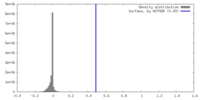
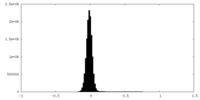
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34058_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
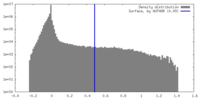
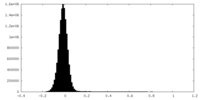
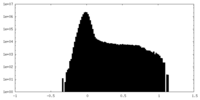
| Density Histograms |
-Half map: #2
| File | emd_34058_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
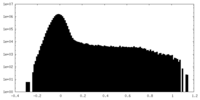
| Density Histograms |
- Sample components
Sample components
-Entire : rO44 A-form (staple xover asymmetry)
| Entire | Name: rO44 A-form (staple xover asymmetry) |
|---|---|
| Components |
|
-Supramolecule #1: rO44 A-form (staple xover asymmetry)
| Supramolecule | Name: rO44 A-form (staple xover asymmetry) / type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Inovirus M13 Inovirus M13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: O (octahedral) / Resolution.type: BY AUTHOR / Resolution: 17.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN2 (ver. 2.3) / Number images used: 22340 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)