+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Csy-AcrIF4-dsDNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IMMUNE SYSTEM-RNA-DNA complex | |||||||||
| Function / homology | CRISPR-associated protein Csy1 / CRISPR-associated protein (Cas_Csy1) / CRISPR-associated protein Csy2 / CRISPR-associated protein (Cas_Csy2) / CRISPR-associated protein Csy3 / CRISPR-associated protein (Cas_Csy3) / Uncharacterized protein / CRISPR-associated protein Csy3 / Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
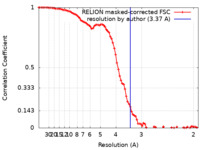
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Feng Y / Zhang LX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage. Authors: Zhengyu Gao / Laixing Zhang / Zihao Ge / Hao Wang / Yourun Yue / Zhuobing Jiang / Xin Wang / Chenying Xu / Yi Zhang / Maojun Yang / Yue Feng /  Abstract: The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system provides prokaryotes with protection against mobile genetic elements such as phages. In turn, phages deploy anti- ...The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system provides prokaryotes with protection against mobile genetic elements such as phages. In turn, phages deploy anti-CRISPR (Acr) proteins to evade this immunity. AcrIF4, an Acr targeting the type I-F CRISPR-Cas system, has been reported to bind the crRNA-guided surveillance (Csy) complex. However, it remains controversial whether AcrIF4 inhibits target DNA binding to the Csy complex. Here, we present structural and mechanistic studies into AcrIF4, exploring its unique anti-CRISPR mechanism. While the Csy-AcrIF4 complex displays decreased affinity for target DNA, it is still able to bind the DNA. Our structural and functional analyses of the Csy-AcrIF4-dsDNA complex revealed that AcrIF4 binding prevents rotation of the helical bundle of the Cas8f subunit induced by dsDNA binding, therefore resulting in failure of nuclease Cas2/3 recruitment and DNA cleavage. Overall, our study provides an interesting example of attack on the nuclease recruitment event by an Acr, but not conventional mechanisms of blocking binding of target DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33837.map.gz emd_33837.map.gz | 44 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33837-v30.xml emd-33837-v30.xml emd-33837.xml emd-33837.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33837_fsc.xml emd_33837_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_33837.png emd_33837.png | 66.8 KB | ||
| Filedesc metadata |  emd-33837.cif.gz emd-33837.cif.gz | 6.8 KB | ||
| Others |  emd_33837_additional_1.map.gz emd_33837_additional_1.map.gz emd_33837_half_map_1.map.gz emd_33837_half_map_1.map.gz emd_33837_half_map_2.map.gz emd_33837_half_map_2.map.gz | 7.2 MB 40.8 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33837 http://ftp.pdbj.org/pub/emdb/structures/EMD-33837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33837 | HTTPS FTP |
-Validation report
| Summary document |  emd_33837_validation.pdf.gz emd_33837_validation.pdf.gz | 797 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33837_full_validation.pdf.gz emd_33837_full_validation.pdf.gz | 796.6 KB | Display | |
| Data in XML |  emd_33837_validation.xml.gz emd_33837_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_33837_validation.cif.gz emd_33837_validation.cif.gz | 19.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33837 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33837 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33837 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33837 | HTTPS FTP |
-Related structure data
| Related structure data |  7yhsMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33837.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33837.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
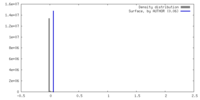
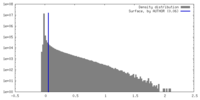
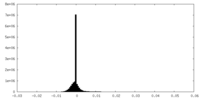
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_33837_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
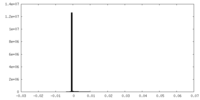
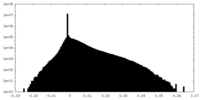
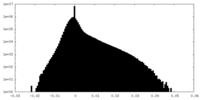
| Density Histograms |
-Half map: #2
| File | emd_33837_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
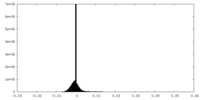
| Density Histograms |
-Half map: #1
| File | emd_33837_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
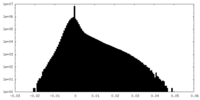
| Density Histograms |
- Sample components
Sample components
+Entire : Csy-AcrIF4-dsDNA
+Supramolecule #1: Csy-AcrIF4-dsDNA
+Supramolecule #2: Csy-AcrIF4
+Supramolecule #3: RNA
+Supramolecule #4: DNA
+Macromolecule #1: Type I-F CRISPR-associated protein Csy1
+Macromolecule #2: CRISPR-associated protein Csy3
+Macromolecule #3: CRISPR type I-F/YPEST-associated protein Csy2
+Macromolecule #4: AcrIF4
+Macromolecule #5: Csy4
+Macromolecule #6: RNA (58-MER)
+Macromolecule #7: DNA (39-MER)
+Macromolecule #8: DNA (5'-D(P*AP*GP*CP*AP*GP*CP*TP*GP*CP*AP*CP*C)-3')
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DARK FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.7 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)