[English] 日本語
 Yorodumi
Yorodumi- EMDB-33650: SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | |||||||||||||||||||||||||||
 Map data Map data | SARS-CoV-2 spike (S) protein trimer in complex with anti-receptor binding domain (RBD) Fab E7 | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | VIRAL PROTEIN | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / membrane fusion / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
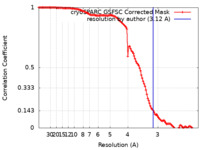
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||||||||||||||||||||
 Authors Authors | Chia WN / Tan CW / Tan AWK / Young B / Starr TN / Lopez E / Fibriansah G / Barr J / Cheng S / Yeoh AYY ...Chia WN / Tan CW / Tan AWK / Young B / Starr TN / Lopez E / Fibriansah G / Barr J / Cheng S / Yeoh AYY / Yap WC / Lim BL / Ng TS / Sia WR / Zhu F / Chen S / Zhang J / Greaney AJ / Chen M / Au GG / Paradkar P / Peiris M / Chung AW / Bloom JD / Lye D / Lok SM / Wang LF | |||||||||||||||||||||||||||
| Funding support |  Singapore, Singapore,  Australia, Australia,  United States, 8 items United States, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor. Authors: Wan Ni Chia / Chee Wah Tan / Aaron Wai Kit Tan / Barnaby Young / Tyler N Starr / Ester Lopez / Guntur Fibriansah / Jennifer Barr / Samuel Cheng / Aileen Ying-Yan Yeoh / Wee Chee Yap / Beng ...Authors: Wan Ni Chia / Chee Wah Tan / Aaron Wai Kit Tan / Barnaby Young / Tyler N Starr / Ester Lopez / Guntur Fibriansah / Jennifer Barr / Samuel Cheng / Aileen Ying-Yan Yeoh / Wee Chee Yap / Beng Lee Lim / Thiam-Seng Ng / Wan Rong Sia / Feng Zhu / Shiwei Chen / Jinyan Zhang / Madeline Sheng Si Kwek / Allison J Greaney / Mark Chen / Gough G Au / Prasad N Paradkar / Malik Peiris / Amy W Chung / Jesse D Bloom / David Lye / Sheemei Lok / Lin-Fa Wang /     Abstract: The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern such as Omicron hampered efforts in controlling the ongoing coronavirus disease 2019 pandemic due to ...The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern such as Omicron hampered efforts in controlling the ongoing coronavirus disease 2019 pandemic due to their ability to escape neutralizing antibodies induced by vaccination or prior infection, highlighting the need to develop broad-spectrum vaccines and therapeutics. Most human monoclonal antibodies (mAbs) reported to date have not demonstrated true pan-sarbecovirus neutralizing breadth especially against animal sarbecoviruses. Here, we report the isolation and characterization of highly potent mAbs targeting the receptor binding domain (RBD) of huACE2-dependent sarbecovirus from a SARS-CoV survivor vaccinated with BNT162b2. Among the six mAbs identified, one (E7) showed better huACE2-dependent sarbecovirus neutralizing potency and breadth than any other mAbs reported to date. Mutagenesis and cryo-electron microscopy studies indicate that these mAbs have a unique RBD contact footprint and that E7 binds to a quaternary structure-dependent epitope. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33650.map.gz emd_33650.map.gz | 161.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33650-v30.xml emd-33650-v30.xml emd-33650.xml emd-33650.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33650_fsc.xml emd_33650_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_33650.png emd_33650.png | 138.8 KB | ||
| Filedesc metadata |  emd-33650.cif.gz emd-33650.cif.gz | 8.3 KB | ||
| Others |  emd_33650_half_map_1.map.gz emd_33650_half_map_1.map.gz emd_33650_half_map_2.map.gz emd_33650_half_map_2.map.gz | 165.4 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33650 http://ftp.pdbj.org/pub/emdb/structures/EMD-33650 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33650 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33650 | HTTPS FTP |
-Validation report
| Summary document |  emd_33650_validation.pdf.gz emd_33650_validation.pdf.gz | 876.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33650_full_validation.pdf.gz emd_33650_full_validation.pdf.gz | 876 KB | Display | |
| Data in XML |  emd_33650_validation.xml.gz emd_33650_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  emd_33650_validation.cif.gz emd_33650_validation.cif.gz | 26.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33650 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33650 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33650 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33650 | HTTPS FTP |
-Related structure data
| Related structure data |  7y71MC  7y72C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33650.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33650.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 spike (S) protein trimer in complex with anti-receptor binding domain (RBD) Fab E7 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.105 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33650_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33650_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment ...
| Entire | Name: SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment ...
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fab E7 light chain
| Macromolecule | Name: Fab E7 light chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.920668 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPLS LPVTPGEPAS ISCRSSQSLL QNNGYNYLAW YLQKPGQSPQ LLIYLSSTRA SGVPDRFSGS GSGTDFTLKI SRVEAEDVG VYYCMQSLQI PGTFGQGTRL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: DIVMTQSPLS LPVTPGEPAS ISCRSSQSLL QNNGYNYLAW YLQKPGQSPQ LLIYLSSTRA SGVPDRFSGS GSGTDFTLKI SRVEAEDVG VYYCMQSLQI PGTFGQGTRL EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Macromolecule #2: Fab E7 heavy chain
| Macromolecule | Name: Fab E7 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.339156 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCTVSGGFIG PHYWSWVRQP PGKGLEWIGY IYISGSTNYN PSLKSRLTIS VDMSKSQFSL TLSSATAAD TAVYYCARGG GYLETGPFEY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS ...String: QVQLQESGPG LVKPSETLSL TCTVSGGFIG PHYWSWVRQP PGKGLEWIGY IYISGSTNYN PSLKSRLTIS VDMSKSQFSL TLSSATAAD TAVYYCARGG GYLETGPFEY WGQGTLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKRV EPK |
-Macromolecule #3: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 132.860156 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HAIHVSGTNG TKRFDNPVLP FNDGVYFAST EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLGVY YHKNNKSWME SEFRVYSSAN NCTFEYVSQP F LMDLEGKQ ...String: VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HAIHVSGTNG TKRFDNPVLP FNDGVYFAST EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLGVY YHKNNKSWME SEFRVYSSAN NCTFEYVSQP F LMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPINLV RDLPQGFSAL EPLVDLPIGI NITRFQTLLA LHRSYLTPGD SS SGWTAGA AAYYVGYLQP RTFLLKYNEN GTITDAVDCA LDPLSETKCT LKSFTVEKGI YQTSNFRVQP TESIVRFPNI TNL CPFGEV FNATRFASVY AWNRKRISNC VADYSVLYNS ASFSTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQT GKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSY GFQP TNGVGYQPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNGLTGT GVLTESNKKF LPFQQFGRDI ADTTDA VRD PQTLEILDIT PCSFGGVSVI TPGTNTSNQV AVLYQDVNCT EVPVAIHADQ LTPTWRVYST GSNVFQTRAG CLIGAEH VN NSYECDIPIG AGICASYQTQ TNSPRAAASV ASQSIIAYTM SLGAENSVAY SNNSIAIPTN FTISVTTEIL PVSMTKTS V DCTMYICGDS TECSNLLLQY GSFCTQLNRA LTGIAVEQDK NTQEVFAQVK QIYKTPPIKD FGGFNFSQIL PDPSKPSKR SPIEDLLFNK VTLADAGFIK QYGDCLGDIA ARDLICAQKF NGLTVLPPLL TDEMIAQYTS ALLAGTITSG WTFGAGPALQ IPFPMQMAY RFNGIGVTQN VLYENQKLIA NQFNSAIGKI QDSLSSTPSA LGKLQDVVNQ NAQALNTLVK QLSSNFGAIS S VLNDILSR LDPPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QS APHGVVF LHVTYVPAQE KNFTTAPAIC HDGKAHFPRE GVFVSNGTHW FVTQRNFYEP QIITTDNTFV SGNCDVVIGI VNN TVYDPL QPELDSFKEE LDKYFKNHTS PDVDLGDISG INASVVNIQK EIDRLNEVAK NLNESLIDLQ ELGKYEQYIK WP UniProtKB: Spike glycoprotein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 14 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 4.76 sec. / Average electron dose: 31.6 e/Å2 Details: Images were collected in counting mode at 25 frames per movie with an exposure time of 4.76 s per movie. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | ||||||||
| Output model |  PDB-7y71: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)