[English] 日本語
 Yorodumi
Yorodumi- EMDB-33105: Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 6.8 Å | |||||||||
 Authors Authors | Wu D / Liu Y / Dai Y / Wang G / Lu G / Chen Y / Li N / Lin J / Gao N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization. Authors: Damu Wu / Yan Liu / Yuhao Dai / Guopeng Wang / Guoliang Lu / Yan Chen / Ningning Li / Jinzhong Lin / Ning Gao /  Abstract: The human AAA+ ATPase CLPB (SKD3) is a protein disaggregase in the mitochondrial intermembrane space (IMS) and functions to promote the solubilization of various mitochondrial proteins. Loss-of- ...The human AAA+ ATPase CLPB (SKD3) is a protein disaggregase in the mitochondrial intermembrane space (IMS) and functions to promote the solubilization of various mitochondrial proteins. Loss-of-function CLPB mutations are associated with a few human diseases with neutropenia and neurological disorders. Unlike canonical AAA+ proteins, CLPB contains a unique ankyrin repeat domain (ANK) at its N-terminus. How CLPB functions as a disaggregase and the role of its ANK domain are currently unclear. Herein, we report a comprehensive structural characterization of human CLPB in both the apo- and substrate-bound states. CLPB assembles into homo-tetradecamers in apo-state and is remodeled into homo-dodecamers upon substrate binding. Conserved pore-loops (PLs) on the ATPase domains form a spiral staircase to grip and translocate the substrate in a step-size of 2 amino acid residues. The ANK domain is not only responsible for maintaining the higher-order assembly but also essential for the disaggregase activity. Interactome analysis suggests that the ANK domain may directly interact with a variety of mitochondrial substrates. These results reveal unique properties of CLPB as a general disaggregase in mitochondria and highlight its potential as a target for the treatment of various mitochondria-related diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33105.map.gz emd_33105.map.gz | 9.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33105-v30.xml emd-33105-v30.xml emd-33105.xml emd-33105.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33105.png emd_33105.png | 56 KB | ||
| Others |  emd_33105_half_map_1.map.gz emd_33105_half_map_1.map.gz emd_33105_half_map_2.map.gz emd_33105_half_map_2.map.gz | 80.7 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33105 http://ftp.pdbj.org/pub/emdb/structures/EMD-33105 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33105 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33105 | HTTPS FTP |
-Validation report
| Summary document |  emd_33105_validation.pdf.gz emd_33105_validation.pdf.gz | 658.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33105_full_validation.pdf.gz emd_33105_full_validation.pdf.gz | 657.6 KB | Display | |
| Data in XML |  emd_33105_validation.xml.gz emd_33105_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_33105_validation.cif.gz emd_33105_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33105 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33105 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33105 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33105 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33105.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33105.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
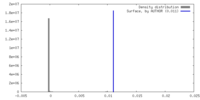
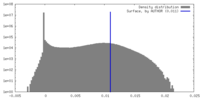
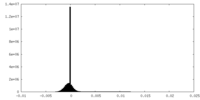
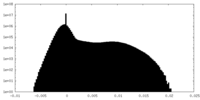
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33105_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
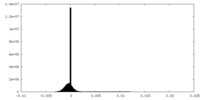
| Density Histograms |
-Half map: #2
| File | emd_33105_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
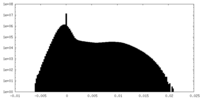
| Density Histograms |
- Sample components
Sample components
-Entire : cryo-EM structure of CLPB in apo-state
| Entire | Name: cryo-EM structure of CLPB in apo-state |
|---|---|
| Components |
|
-Supramolecule #1: cryo-EM structure of CLPB in apo-state
| Supramolecule | Name: cryo-EM structure of CLPB in apo-state / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: CLPB
| Macromolecule | Name: CLPB / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MLGSLVLRRK ALAPRLLLRL LRSPTLRGHG GASGRNVTTG SLGEPQWLRV ATGGRPGTSP ALFSGRGAA TGGRQGGRFD TKCLAAATWG RLPGPEETLP GQDSWNGVPS RAGLGMCALA A ALVVHCYS KSPSNKDAAL LEAARANNMQ EVSRLLSEGA DVNAKHRLGW ...String: MLGSLVLRRK ALAPRLLLRL LRSPTLRGHG GASGRNVTTG SLGEPQWLRV ATGGRPGTSP ALFSGRGAA TGGRQGGRFD TKCLAAATWG RLPGPEETLP GQDSWNGVPS RAGLGMCALA A ALVVHCYS KSPSNKDAAL LEAARANNMQ EVSRLLSEGA DVNAKHRLGW TALMVAAINR NN SVVQVLL AAGADPNLGD DFSSVYKTAK EQGIHSLEVL ITREDDFNNR LNNRASFKGC TAL HYAVLA DDYRTVKELL DGGANPLQRN EMGHTPLDYA REGEVMKLLR TSEAKYQEKQ RKRE AEERR RFPLEQRLKE HIIGQESAIA TVGAAIRRKE NGWYDEEHPL VFLFLGSSGI GKTEL AKQT AKYMHKDAKK GFIRLDMSEF QERHEVAKFI GSPPGYVGHE EGGQLTKKLK QCPNAV VLF DEVDKAHPDV LTIMLQLFDE GRLTDGKGKT IDCKDAIFIM TSNVASDEIA QHALQLR QE ALEMSRNRIA ENLGDVQISD KITISKNFKE NVIRPILKAH FRRDEFLGRI NEIVYFLP F CHSELIQLVN KELNFWAKRA KQRHNITLLW DREVADVLVD GYNVHYGARS IKHEVERRV VNQLAAAYEQ DLLPGGCTLR ITVEDSDKQL LKSPELPSPQ AEKRLPKLRL EIIDKDSKTR RLDIRAPLH PEKVCNTI |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DARK FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 153005 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)