[English] 日本語
 Yorodumi
Yorodumi- EMDB-3286: Evidence for a conformational switch in Influenzavirus M1 and its... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3286 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Evidence for a conformational switch in Influenzavirus M1 and its role in filamentous virion architecture | |||||||||
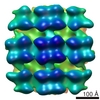
 Map data Map data | Reconstruction of inner membrane leaflet, M1 and attached densities at leading virion tips | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza A virus / FLUAV / polymerase / M1 / crown / leading tip / trailing tip / filamentous | |||||||||
| Biological species |   Influenza A virus Influenza A virus | |||||||||
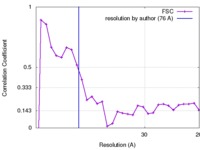
| Method | subtomogram averaging / cryo EM / Resolution: 76.0 Å | |||||||||
 Authors Authors | Kiss G / Abdulsattar BO / Phapugrangkul P / Birch K / Jones IM / Neuman BW | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Evidence for a conformational switch in Influenzavirus M1 and its role in filamentous virion architecture Authors: Kiss G / Abdulsattar BO / Phapugrangkul P / Birch K / Jones IM / Neuman BW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3286.map.gz emd_3286.map.gz | 923.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3286-v30.xml emd-3286-v30.xml emd-3286.xml emd-3286.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3286_fsc.xml emd_3286_fsc.xml | 2.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3286.png emd_3286.png | 84.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3286 http://ftp.pdbj.org/pub/emdb/structures/EMD-3286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3286 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3286 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3286.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3286.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of inner membrane leaflet, M1 and attached densities at leading virion tips | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
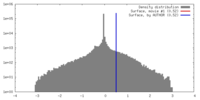
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Leading tip of filamentous Influenza A virions including inner le...
| Entire | Name: Leading tip of filamentous Influenza A virions including inner leaflet, M1 and internal crown |
|---|---|
| Components |
|
-Supramolecule #1000: Leading tip of filamentous Influenza A virions including inner le...
| Supramolecule | Name: Leading tip of filamentous Influenza A virions including inner leaflet, M1 and internal crown type: sample / ID: 1000 Details: Compare to matched image of the opposite virion tip Number unique components: 1 |
|---|
-Supramolecule #1: Influenza A virus
| Supramolecule | Name: Influenza A virus / type: virus / ID: 1 / Name.synonym: Influenza virus Details: Reconstruction of the inner layer of the viral envelope and attached densities likely representing polymerase molecules from native virions of mixed A/Aichi/X31 and A/Udorn strains. Map is ...Details: Reconstruction of the inner layer of the viral envelope and attached densities likely representing polymerase molecules from native virions of mixed A/Aichi/X31 and A/Udorn strains. Map is Gaussian filtered to 7.6 nm the 0.5 FSC. Reconstructed in EMAN2. NCBI-ID: 11320 / Sci species name: Influenza A virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No / Syn species name: Influenza virus |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / Strain: Combined A/Aichi/2/68 and A/Udorn/307/72 / synonym: VERTEBRATES Homo sapiens (human) / Strain: Combined A/Aichi/2/68 and A/Udorn/307/72 / synonym: VERTEBRATES |
| Host system | Organism:  Homo sapiens (human) / Recombinant cell: MDCK Homo sapiens (human) / Recombinant cell: MDCK |
| Virus shell | Shell ID: 1 / Name: M1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK III Details: Details for EM and reconstruction published in Calder et al., Proc Natl Acad Sci U S A. 2010 Jun 8; 107(23): 10685 to 10690. Method: Blotted 4s before plunging |
|---|
- Electron microscopy
Electron microscopy
| Microscope | OTHER |
|---|---|
| Date | Jan 1, 2010 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (2k x 2k) / Average electron dose: 50 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera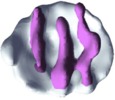





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)