+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM map of PRP22 region of human pre-C*-I complex | |||||||||
 Map data Map data | The PRP22 region of human pre-C*-I complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Zhan X / Lu Y / Shi Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Mechanism of exon ligation by human spliceosome. Authors: Xiechao Zhan / Yichen Lu / Xiaofeng Zhang / Chuangye Yan / Yigong Shi /  Abstract: Pre-mRNA splicing involves two sequential reactions: branching and exon ligation. The C complex after branching undergoes remodeling to become the C complex, which executes exon ligation. Here, we ...Pre-mRNA splicing involves two sequential reactions: branching and exon ligation. The C complex after branching undergoes remodeling to become the C complex, which executes exon ligation. Here, we report cryo-EM structures of two intermediate human spliceosomal complexes, pre-C-I and pre-C-II, both at 3.6 Å. In both structures, the 3' splice site is already docked into the active site, the ensuing 3' exon sequences are anchored on PRP8, and the step II factor FAM192A contacts the duplex between U2 snRNA and the branch site. In the transition of pre-C-I to pre-C-II, the step II factors Cactin, FAM32A, PRKRIP1, and SLU7 are recruited. Notably, the RNA helicase PRP22 is positioned quite differently in the pre-C-I, pre-C-II, and C complexes, suggesting a role in 3' exon binding and proofreading. Together with information on human C and C complexes, our studies recapitulate a molecular choreography of the C-to-C transition, revealing mechanistic insights into exon ligation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32318.map.gz emd_32318.map.gz | 122.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32318-v30.xml emd-32318-v30.xml emd-32318.xml emd-32318.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32318.png emd_32318.png | 50.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32318 http://ftp.pdbj.org/pub/emdb/structures/EMD-32318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32318 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32318 | HTTPS FTP |
-Validation report
| Summary document |  emd_32318_validation.pdf.gz emd_32318_validation.pdf.gz | 433.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32318_full_validation.pdf.gz emd_32318_full_validation.pdf.gz | 433 KB | Display | |
| Data in XML |  emd_32318_validation.xml.gz emd_32318_validation.xml.gz | 7.4 KB | Display | |
| Data in CIF |  emd_32318_validation.cif.gz emd_32318_validation.cif.gz | 8.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32318 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32318 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32318 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32318 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32318.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32318.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The PRP22 region of human pre-C*-I complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.338 Å | ||||||||||||||||||||||||||||||||||||
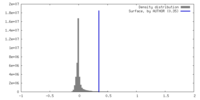
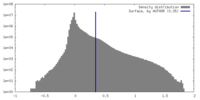
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Human pre-C*-I complex
| Entire | Name: Human pre-C*-I complex |
|---|---|
| Components |
|
-Supramolecule #1: Human pre-C*-I complex
| Supramolecule | Name: Human pre-C*-I complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#43 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 68202 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)