[English] 日本語
 Yorodumi
Yorodumi- EMDB-32111: Cryo-EM structure of a human ATP11C-CDC50A flippase reconstituted... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32111 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a human ATP11C-CDC50A flippase reconstituted in the Nanodisc in E1P state. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of phospholipid translocation / aminophospholipid flippase activity / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex / ATPase-coupled intramembrane lipid transporter activity / phosphatidylserine floppase activity / positive regulation of protein exit from endoplasmic reticulum / xenobiotic transmembrane transport ...positive regulation of phospholipid translocation / aminophospholipid flippase activity / aminophospholipid transport / phosphatidylserine flippase activity / protein localization to endosome / phospholipid-translocating ATPase complex / ATPase-coupled intramembrane lipid transporter activity / phosphatidylserine floppase activity / positive regulation of protein exit from endoplasmic reticulum / xenobiotic transmembrane transport / phosphatidylethanolamine flippase activity / P-type phospholipid transporter / phospholipid translocation / transport vesicle membrane / azurophil granule membrane / Ion transport by P-type ATPases / specific granule membrane / monoatomic ion transmembrane transport / recycling endosome / positive regulation of neuron projection development / recycling endosome membrane / late endosome membrane / early endosome membrane / apical plasma membrane / lysosomal membrane / Neutrophil degranulation / endoplasmic reticulum membrane / structural molecule activity / Golgi apparatus / magnesium ion binding / endoplasmic reticulum / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
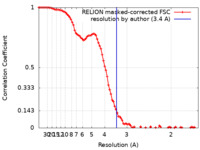
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Nakanishii H / Abe K | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2022 Journal: J Biol Chem / Year: 2022Title: Cryo-EM of the ATP11C flippase reconstituted in Nanodiscs shows a distended phospholipid bilayer inner membrane around transmembrane helix 2. Authors: Hanayo Nakanishi / Kenichi Hayashida / Tomohiro Nishizawa / Atsunori Oshima / Kazuhiro Abe /  Abstract: ATP11C is a member of the P4-ATPase flippase family that mediates translocation of phosphatidylserine (PtdSer) across the lipid bilayer. In order to characterize the structure and function of ATP11C ...ATP11C is a member of the P4-ATPase flippase family that mediates translocation of phosphatidylserine (PtdSer) across the lipid bilayer. In order to characterize the structure and function of ATP11C in a model natural lipid environment, we revisited and optimized a quick procedure for reconstituting ATP11C into Nanodiscs using methyl-β-cyclodextrin as a reagent for the detergent removal. ATP11C was efficiently reconstituted with the endogenous lipid, or the mixture of endogenous lipid and synthetic dioleoylphosphatidylcholine (DOPC)/dioleoylphosphatidylserine (DOPS), all of which retained the ATPase activity. We obtained 3.4 Å and 3.9 Å structures using single-particle cryo-electron microscopy (cryo-EM) of AlF- and BeF-stabilized ATP11C transport intermediates, respectively, in a bilayer containing DOPS. We show that the latter exhibited a distended inner membrane around ATP11C transmembrane helix 2, possibly reflecting the perturbation needed for phospholipid release to the lipid bilayer. Our structures of ATP11C in the lipid membrane indicate that the membrane boundary varies upon conformational changes of the enzyme and is no longer flat around the protein, a change that likely contributes to phospholipid translocation across the membrane leaflets. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
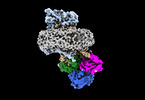
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32111.map.gz emd_32111.map.gz | 8.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32111-v30.xml emd-32111-v30.xml emd-32111.xml emd-32111.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32111_fsc.xml emd_32111_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_32111.png emd_32111.png | 52.8 KB | ||
| Masks |  emd_32111_msk_1.map emd_32111_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_32111_half_map_1.map.gz emd_32111_half_map_1.map.gz emd_32111_half_map_2.map.gz emd_32111_half_map_2.map.gz | 49.5 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32111 http://ftp.pdbj.org/pub/emdb/structures/EMD-32111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32111 | HTTPS FTP |
-Validation report
| Summary document |  emd_32111_validation.pdf.gz emd_32111_validation.pdf.gz | 732.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32111_full_validation.pdf.gz emd_32111_full_validation.pdf.gz | 732.4 KB | Display | |
| Data in XML |  emd_32111_validation.xml.gz emd_32111_validation.xml.gz | 15.4 KB | Display | |
| Data in CIF |  emd_32111_validation.cif.gz emd_32111_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32111 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32111 | HTTPS FTP |
-Related structure data
| Related structure data |  7vshMC  7vsgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32111.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32111.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32111_msk_1.map emd_32111_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32111_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_32111_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of a human ATP11C-CDC50A flippase reconstituted...
| Entire | Name: Cryo-EM structure of a human ATP11C-CDC50A flippase reconstituted in the Nanodisc in E1P state. |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of a human ATP11C-CDC50A flippase reconstituted...
| Supramolecule | Name: Cryo-EM structure of a human ATP11C-CDC50A flippase reconstituted in the Nanodisc in E1P state. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: Mammalia (mammals) |
-Macromolecule #1: Phospholipid-transporting ATPase IG
| Macromolecule | Name: Phospholipid-transporting ATPase IG / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: P-type phospholipid transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.403617 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: RFCAGEEKRV GTRTVFVGNH PVSETEAYIA QRFCDNRIVS SKYTLWNFLP KNLFEQFRRI ANFYFLIIFL VQVTVDTPTS PVTSGLPLF FVITVTAIKQ GYEDWLRHRA DNEVNKSTVY IIENAKRVRK ESEKIKVGDV VEVQADETFP CDLILLSSCT T DGTCYVTT ...String: RFCAGEEKRV GTRTVFVGNH PVSETEAYIA QRFCDNRIVS SKYTLWNFLP KNLFEQFRRI ANFYFLIIFL VQVTVDTPTS PVTSGLPLF FVITVTAIKQ GYEDWLRHRA DNEVNKSTVY IIENAKRVRK ESEKIKVGDV VEVQADETFP CDLILLSSCT T DGTCYVTT ASLDGESNCK THYAVRDTIA LCTAESIDTL RAAIECEQPQ PDLYKFVGRI NIYSNSLEAV ARSLGPENLL LK GATLKNT EKIYGVAVYT GMETKMALNY QGKSQKRSAV EKSINAFLIV YLFILLTKAA VCTTLKYVWQ STPYNDEPWY NQK TQKERE TLKVLKMFTD FLSFMVLFNF IIPVSMYVTV EMQKFLGSFF ISWDKDFYDE EINEGALVNT SDLNEELGQV DYVF TDKTG TLTENSMEFI ECCIDGHKYK GVTQEVDGLS QTDGTLTYFD KVDKNREELF LRALCLCHTV EIKTNDAVDG ATESA ELTY ISSSPDEIAL VKGAKRYGFT FLGNRNGYMR VENQRKEIEE YELLHTLNFD AVRRRMSVIV KTQEGDILLF CKGADS AVF PRVQNHEIEL TKVHVERNAM DGYRTLCVAF KEIAPDDYER INRQLIEAKM ALQDREEKME KVFDDIETNM NLIGATA VE DKLQDQAAET IEALHAAGLK VWVLTGDKME TAKSTCYACR LFQTNTELLE LTTKTIEESE RKEDRLHELL IEYRKKLL H EFPKSTRSFK KAWTEHQEYG LIIDGSTLSL ILNSSQDSSS NNYKSIFLQI CMKCTAVLCC RMAPLQKAQI VRMVKNLKG SPITLSIGDG ANDVSMILES HVGIGIKGKE GRQAARNSDY SVPKFKHLKK LLLAHGHLYY VRIAHLVQYF FYKNLCFILP QFLYQFFCG FSQQPLYDAA YLTMYNICFT SLPILAYSLL EQHINIDTLT SDPRLYMKIS GNAMLQLGPF LYWTFLAAFE G TVFFFGTY FLFQTASLEE NGKVYGNWTF GTIVFTVLVF TVTLKLALDT RFWTWINHFV IWGSLAFYVF FSFFWGGIIW PF LKQQRMY FVFAQMLSSV STWLAIILLI FISLFPEILL IVLKNVR |
-Macromolecule #2: Cell cycle control protein 50A
| Macromolecule | Name: Cell cycle control protein 50A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.727527 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: MAMNYNAKDE VDGGPPCAPG GTAKTRRPDN TAFKQQRLPA WQPILTAGTV LPIFFIIGLI FIPIGIGIFV TSNNIREIEI DYTGTEPSS PCNKCLSPDV TPCFCTINFT LEKSFEGNVF MYYGLSNFYQ NHRRYVKSRD DSQLNGDSSA LLNPSKECEP Y RRNEDKPI ...String: MAMNYNAKDE VDGGPPCAPG GTAKTRRPDN TAFKQQRLPA WQPILTAGTV LPIFFIIGLI FIPIGIGIFV TSNNIREIEI DYTGTEPSS PCNKCLSPDV TPCFCTINFT LEKSFEGNVF MYYGLSNFYQ NHRRYVKSRD DSQLNGDSSA LLNPSKECEP Y RRNEDKPI APCGAIANSM FNDTLELFLI GNDSYPIPIA LKKKGIAWWT DKNVKFRNPP GGDNLEERFK GTTKPVNWLK PV YMLDSDP DNNGFINEDF IVWMRTAALP TFRKLYRLIE RKSDLHPTLP AGRYSLNVTY NYPVHYFDGR KRMILSTISW MGG KNPFLG IAYIAVGSIS FLLGVVLLVI NHKYRNSSNT ADITI |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: TETRAFLUOROALUMINATE ION
| Macromolecule | Name: TETRAFLUOROALUMINATE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: ALF |
|---|---|
| Molecular weight | Theoretical: 102.975 Da |
| Chemical component information |  ChemComp-ALF: |
-Macromolecule #6: O-[(S)-({(2R)-2,3-bis[(9Z)-octadec-9-enoyloxy]propyl}oxy)(hydroxy...
| Macromolecule | Name: O-[(S)-({(2R)-2,3-bis[(9Z)-octadec-9-enoyloxy]propyl}oxy)(hydroxy)phosphoryl]-L-serine type: ligand / ID: 6 / Number of copies: 1 / Formula: 17F |
|---|---|
| Molecular weight | Theoretical: 788.043 Da |
| Chemical component information |  ChemComp-17F: |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.5 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z
Z Y
Y X
X