[English] 日本語
 Yorodumi
Yorodumi- EMDB-30218: Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransfe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30218 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Mycobacterium tuberculosis / cell wall synthesis / drug target / ethambutol / arabinosyltransferase / EmbA / EmbB / EmbC / acyl carrier protein / arabinogalactan / lipoarabinomannan / drug resistance / TRANSFERASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationarabinosyltransferase activity / Actinobacterium-type cell wall biogenesis / lipid A biosynthetic process / acyl binding / acyl carrier activity / Transferases; Glycosyltransferases; Pentosyltransferases / peptidoglycan-based cell wall / cell wall organization / response to antibiotic / membrane ...arabinosyltransferase activity / Actinobacterium-type cell wall biogenesis / lipid A biosynthetic process / acyl binding / acyl carrier activity / Transferases; Glycosyltransferases; Pentosyltransferases / peptidoglycan-based cell wall / cell wall organization / response to antibiotic / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) / Mycobacterium tuberculosis H37Rv (bacteria) /  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||||||||
 Authors Authors | Zhang L / Zhao Y | |||||||||||||||
| Funding support |  China, China,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Authors: Lu Zhang / Yao Zhao / Yan Gao / Lijie Wu / Ruogu Gao / Qi Zhang / Yinan Wang / Chengyao Wu / Fangyu Wu / Sudagar S Gurcha / Natacha Veerapen / Sarah M Batt / Wei Zhao / Ling Qin / Xiuna Yang ...Authors: Lu Zhang / Yao Zhao / Yan Gao / Lijie Wu / Ruogu Gao / Qi Zhang / Yinan Wang / Chengyao Wu / Fangyu Wu / Sudagar S Gurcha / Natacha Veerapen / Sarah M Batt / Wei Zhao / Ling Qin / Xiuna Yang / Manfu Wang / Yan Zhu / Bing Zhang / Lijun Bi / Xian'en Zhang / Haitao Yang / Luke W Guddat / Wenqing Xu / Quan Wang / Jun Li / Gurdyal S Besra / Zihe Rao /    Abstract: The arabinosyltransferases EmbA, EmbB, and EmbC are involved in cell wall synthesis and are recognized as targets for the anti-tuberculosis drug ethambutol. In this study, we determined cryo- ...The arabinosyltransferases EmbA, EmbB, and EmbC are involved in cell wall synthesis and are recognized as targets for the anti-tuberculosis drug ethambutol. In this study, we determined cryo-electron microscopy and x-ray crystal structures of mycobacterial EmbA-EmbB and EmbC-EmbC complexes in the presence of their glycosyl donor and acceptor substrates and with ethambutol. These structures show how the donor and acceptor substrates bind in the active site and how ethambutol inhibits arabinosyltransferases by binding to the same site as both substrates in EmbB and EmbC. Most drug-resistant mutations are located near the ethambutol binding site. Collectively, our work provides a structural basis for understanding the biochemical function and inhibition of arabinosyltransferases and the development of new anti-tuberculosis agents. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30218.map.gz emd_30218.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30218-v30.xml emd-30218-v30.xml emd-30218.xml emd-30218.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30218.png emd_30218.png | 67 KB | ||
| Masks |  emd_30218_msk_1.map emd_30218_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-30218.cif.gz emd-30218.cif.gz | 7.8 KB | ||
| Others |  emd_30218_half_map_1.map.gz emd_30218_half_map_1.map.gz emd_30218_half_map_2.map.gz emd_30218_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30218 http://ftp.pdbj.org/pub/emdb/structures/EMD-30218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30218 | HTTPS FTP |
-Related structure data
| Related structure data |  7bvfMC  7bvcC  7bveC  7bvgC  7bvhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30218.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30218.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_30218_msk_1.map emd_30218_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
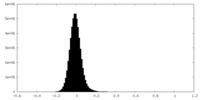
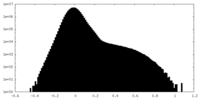
| Density Histograms |
-Half map: #1
| File | emd_30218_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_30218_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 ...
+Supramolecule #1: Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 ...
+Supramolecule #2: EmbA and EmbB
+Supramolecule #3: AcpM
+Macromolecule #1: Probable arabinosyltransferase B
+Macromolecule #2: Probable arabinosyltransferase A
+Macromolecule #3: Meromycolate extension acyl carrier protein
+Macromolecule #4: CALCIUM ION
+Macromolecule #5: Ethambutol
+Macromolecule #6: MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE
+Macromolecule #7: CARDIOLIPIN
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 101.325 kPa | ||||||||
| Vitrification | Cryogen name: ETHANE | ||||||||
| Details | The sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 78.5 K / Max: 78.6 K |
| Alignment procedure | Coma free - Residual tilt: 10.0 mrad |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Number grids imaged: 1 / #0 - Number real images: 7612 / #0 - Average exposure time: 2.0 sec. / #0 - Average electron dose: 50.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Average electron dose: 50.0 e/Å2 / #2 - Image recording ID: 3 / #2 - Film or detector model: GATAN K3 (6k x 4k) / #2 - Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7bvf: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)