+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of AAV2-R404A Variant | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Symmetry / capsid / AAV2 / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont entry into host cell via permeabilization of host membrane / host cell nucleolus / T=1 icosahedral viral capsid / clathrin-dependent endocytosis of virus by host cell / virion attachment to host cell / structural molecule activity Similarity search - Function | |||||||||
| Biological species |   Adeno-associated virus / Adeno-associated virus /  adeno-associated virus 2 adeno-associated virus 2 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.62 Å | |||||||||
 Authors Authors | Bennett AD / McKenna R | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: to be published Journal: to be publishedTitle: Cryo-EM Structure of genome containing AAV2 Authors: Bennett AD / Mckenna R | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29636.map.gz emd_29636.map.gz | 226.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29636-v30.xml emd-29636-v30.xml emd-29636.xml emd-29636.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29636.png emd_29636.png | 125.6 KB | ||
| Filedesc metadata |  emd-29636.cif.gz emd-29636.cif.gz | 5.8 KB | ||
| Others |  emd_29636_half_map_1.map.gz emd_29636_half_map_1.map.gz emd_29636_half_map_2.map.gz emd_29636_half_map_2.map.gz | 60.8 MB 60.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29636 http://ftp.pdbj.org/pub/emdb/structures/EMD-29636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29636 | HTTPS FTP |
-Validation report
| Summary document |  emd_29636_validation.pdf.gz emd_29636_validation.pdf.gz | 1008.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29636_full_validation.pdf.gz emd_29636_full_validation.pdf.gz | 1007.9 KB | Display | |
| Data in XML |  emd_29636_validation.xml.gz emd_29636_validation.xml.gz | 16.6 KB | Display | |
| Data in CIF |  emd_29636_validation.cif.gz emd_29636_validation.cif.gz | 19.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29636 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29636 | HTTPS FTP |
-Related structure data
| Related structure data |  8fznMC  8fywC  8fz0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29636.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29636.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.962 Å | ||||||||||||||||||||||||||||||||||||
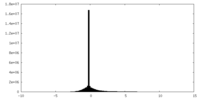
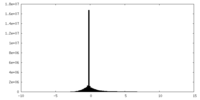
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29636_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
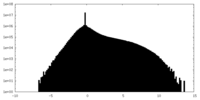
| Density Histograms |
-Half map: #1
| File | emd_29636_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
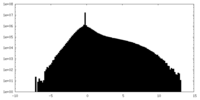
| Density Histograms |
- Sample components
Sample components
-Entire : adeno-associated virus 2
| Entire | Name:  adeno-associated virus 2 adeno-associated virus 2 |
|---|---|
| Components |
|
-Supramolecule #1: adeno-associated virus 2
| Supramolecule | Name: adeno-associated virus 2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10804 / Sci species name: adeno-associated virus 2 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.9 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 26.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Adeno-associated virus Adeno-associated virus |
| Molecular weight | Theoretical: 56.722445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GDRVITTSTR TWALPTYNNH LYKQISSQSG ASNDNHYFGY STPWGYFDFN RFHCHFSPRD WQRLINNNWG FRPKRLNFKL FNIQVKEVT QNDGTTTIAN NLTSTVQVFT DSEYQLPYVL GSAHQGCLPP FPADVFMVPQ YGYLTLNNGS QAVGRSSFYC L EYFPSQML ...String: GDRVITTSTR TWALPTYNNH LYKQISSQSG ASNDNHYFGY STPWGYFDFN RFHCHFSPRD WQRLINNNWG FRPKRLNFKL FNIQVKEVT QNDGTTTIAN NLTSTVQVFT DSEYQLPYVL GSAHQGCLPP FPADVFMVPQ YGYLTLNNGS QAVGRSSFYC L EYFPSQML ATGNNFTFSY TFEDVPFHSS YAHSQSLDRL MNPLIDQYLY YLSRTNTPSG TTTQSRLQFS QAGASDIRDQ SR NWLPGPC YRQQRVSKTS ADNNNSEYSW TGATKYHLNG RDSLVNPGPA MASHKDDEEK FFPQSGVLIF GKQGSEKTNV DIE KVMITD EEEIRTTNPV ATEQYGSVST NLQRGNRQAA TADVNTQGVL PGMVWQDRDV YLQGPIWAKI PHTDGHFHPS PLMG GFGLK HPPPQILIKN TPVPANPSTT FSAAKFASFI TQYSTGQVSV EIEWELQKEN SKRWNPEIQY TSNYNKSVNV DFTVD TNGV YSEPRPIGTR YLTRNL UniProtKB: Capsid protein VP1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.3 / Component - Concentration: 1.0 mM / Component - Formula: PBS-MK / Component - Name: TD / Details: 1XPBS, 1 mM MgCl2, 2.5 mM KCl pH7.3 |
| Grid | Model: Quantifoil / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-64 (8k x 8k) / Number real images: 1674 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.62 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 23783 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8fzn: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)