+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human PTH1R in complex with LA-PTH and Gs | |||||||||||||||
 Map data Map data | sharpened consensus map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | GPCR / agonist / hormone / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationparathyroid hormone receptor activity / G protein-coupled peptide receptor activity / Class B/2 (Secretin family receptors) / osteoblast development / positive regulation of inositol phosphate biosynthetic process / bone mineralization / PKA activation in glucagon signalling / peptide hormone binding / hair follicle placode formation / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger ...parathyroid hormone receptor activity / G protein-coupled peptide receptor activity / Class B/2 (Secretin family receptors) / osteoblast development / positive regulation of inositol phosphate biosynthetic process / bone mineralization / PKA activation in glucagon signalling / peptide hormone binding / hair follicle placode formation / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / developmental growth / D1 dopamine receptor binding / intracellular transport / renal water homeostasis / Hedgehog 'off' state / chondrocyte differentiation / bone resorption / adenylate cyclase-activating adrenergic receptor signaling pathway / activation of adenylate cyclase activity / cell maturation / cellular response to glucagon stimulus / adenylate cyclase activator activity / regulation of insulin secretion / trans-Golgi network membrane / skeletal system development / negative regulation of inflammatory response to antigenic stimulus / bone development / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G protein activity / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / platelet aggregation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / cognition / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / intracellular calcium ion homeostasis / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / sensory perception of smell / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / positive regulation of cold-induced thermogenesis / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (s) signalling events / basolateral plasma membrane / G alpha (q) signalling events / in utero embryonic development / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / receptor complex / cell surface receptor signaling pathway / G protein-coupled receptor signaling pathway / apical plasma membrane / negative regulation of cell population proliferation / lysosomal membrane / GTPase activity / positive regulation of cell population proliferation / synapse / protein-containing complex binding / GTP binding / signal transduction Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||
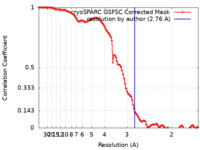
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||||||||
 Authors Authors | Cary BP / Belousoff MJ / Piper SJ / Wootten D / Sexton PM | |||||||||||||||
| Funding support |  Australia, 4 items Australia, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Molecular insights into peptide agonist engagement with the PTH receptor. Authors: Brian P Cary / Elliot J Gerrard / Matthew J Belousoff / Madeleine M Fletcher / Yan Jiang / Isabella C Russell / Sarah J Piper / Denise Wootten / Patrick M Sexton /  Abstract: The parathyroid hormone (PTH) 1 receptor (PTH1R) is a G protein-coupled receptor (GPCR) that regulates skeletal development and calcium homeostasis. Here, we describe cryo-EM structures of the PTH1R ...The parathyroid hormone (PTH) 1 receptor (PTH1R) is a G protein-coupled receptor (GPCR) that regulates skeletal development and calcium homeostasis. Here, we describe cryo-EM structures of the PTH1R in complex with fragments of the two hormones, PTH and PTH-related protein, the drug abaloparatide, as well as the engineered tool compounds, long-acting PTH (LA-PTH) and the truncated peptide, M-PTH(1-14). We found that the critical N terminus of each agonist engages the transmembrane bundle in a topologically similar fashion, reflecting similarities in measures of Gαs activation. The full-length peptides induce subtly different extracellular domain (ECD) orientations relative to the transmembrane domain. In the structure bound to M-PTH, the ECD is unresolved, demonstrating that the ECD is highly dynamic when unconstrained by a peptide. High resolutions enabled identification of water molecules near peptide and G protein binding sites. Our results illuminate the action of orthosteric agonists of the PTH1R. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29287.map.gz emd_29287.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29287-v30.xml emd-29287-v30.xml emd-29287.xml emd-29287.xml | 30.2 KB 30.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29287_fsc.xml emd_29287_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29287.png emd_29287.png | 77.4 KB | ||
| Masks |  emd_29287_msk_1.map emd_29287_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29287.cif.gz emd-29287.cif.gz | 7.6 KB | ||
| Others |  emd_29287_additional_1.map.gz emd_29287_additional_1.map.gz emd_29287_additional_2.map.gz emd_29287_additional_2.map.gz emd_29287_half_map_1.map.gz emd_29287_half_map_1.map.gz emd_29287_half_map_2.map.gz emd_29287_half_map_2.map.gz | 62.2 MB 62.4 MB 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29287 http://ftp.pdbj.org/pub/emdb/structures/EMD-29287 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29287 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29287 | HTTPS FTP |
-Validation report
| Summary document |  emd_29287_validation.pdf.gz emd_29287_validation.pdf.gz | 941.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29287_full_validation.pdf.gz emd_29287_full_validation.pdf.gz | 941.5 KB | Display | |
| Data in XML |  emd_29287_validation.xml.gz emd_29287_validation.xml.gz | 19.2 KB | Display | |
| Data in CIF |  emd_29287_validation.cif.gz emd_29287_validation.cif.gz | 24.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29287 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29287 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29287 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29287 | HTTPS FTP |
-Related structure data
| Related structure data |  8fluMC  8flqC  8flrC  8flsC  8fltC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29287.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29287.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened consensus map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
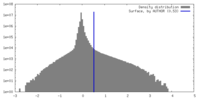
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29287_msk_1.map emd_29287_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened focused (receptor) refinement
| File | emd_29287_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened focused (receptor) refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened consensus map
| File | emd_29287_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened consensus map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_29287_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_29287_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human PTH1R in complex with LA-PTH and Gs
| Entire | Name: Human PTH1R in complex with LA-PTH and Gs |
|---|---|
| Components |
|
-Supramolecule #1: Human PTH1R in complex with LA-PTH and Gs
| Supramolecule | Name: Human PTH1R in complex with LA-PTH and Gs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.683434 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.413863 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: QSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: QSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody35
| Macromolecule | Name: Nanobody35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.885439 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSS |
-Macromolecule #5: Long-acting PTH
| Macromolecule | Name: Long-acting PTH / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.324043 KDa |
| Sequence | String: AVAEIQLMHQ RAKWIQDARR RAFLHKLIAE IHTAEY |
-Macromolecule #6: Parathyroid hormone/parathyroid hormone-related peptide receptor
| Macromolecule | Name: Parathyroid hormone/parathyroid hormone-related peptide receptor type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.513758 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PADDVMTKEE QIFLLHRAQA QCEKRLKEVL QRPASIMESD KGWTSASTSG KPRKDKASG KLYPESEEDK EAPTGSRYRG RPCLPEWDHI LCWPLGAPGE VVAVPCPDYI YDFNHKGHAY RRCDRNGSWE L VPGHNRTW ...String: MKTIIALSYI FCLVFADYKD DDDLEVLFQG PADDVMTKEE QIFLLHRAQA QCEKRLKEVL QRPASIMESD KGWTSASTSG KPRKDKASG KLYPESEEDK EAPTGSRYRG RPCLPEWDHI LCWPLGAPGE VVAVPCPDYI YDFNHKGHAY RRCDRNGSWE L VPGHNRTW ANYSECVKFL TNETREREVF DRLGMIYTVG YSVSLASLTV AVLILAYFRR LHCTRNYIHM HLFLSFMLRA VS IFVKDAV LYSGATLDEA ERLTEEELRA IAQAPPPPAT AAAGYAGCRV AVTFFLYFLA TNYYWILVEG LYLHSLIFMA FFS EKKYLW GFTVFGWGLP AVFVAVWVSV RATLANTGCW DLSSGNKKWI IQVPILASIV LNFILFINIV RVLATKLRET NAGR CDTRQ QYRKLLKSTL VLMPLFGVHY IVFMATPYTE VSGTLWQVQM HYEMLFNSFQ GFFVAIIYCF CNGEVQAEIK KSWSR WTLA LDFKRKARSG SSSYSYGPMV SHTSVTNVGP RVGLGLPLSP RLLPTATTNG HPQLPGHAKP GTPALETLET TPPAMA APK DDGFLNGSCS GLDEEASGPE RPPALLQEEW ETVMPAGLEV LFQGPHHHHH HHH UniProtKB: Parathyroid hormone/parathyroid hormone-related peptide receptor |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 5 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.51 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Buffer also contained 2 micromolar LA-PTH peptide. | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 20 mA current, negative polarity | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 5172 / Average exposure time: 5.03 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)