[English] 日本語
 Yorodumi
Yorodumi- EMDB-29029: Langya Virus Fusion Protein (LayV-F) in Pre-Fusion Conformation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Langya Virus Fusion Protein (LayV-F) in Pre-Fusion Conformation | |||||||||
 Map data Map data | Final 3D refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycoprotein / Fusion / Paramyxovirus / Henipavirus / Viral Protein | |||||||||
| Biological species |  Langya virus Langya virus | |||||||||
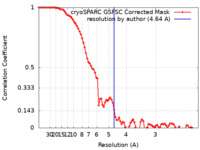
| Method | single particle reconstruction / cryo EM / Resolution: 4.64 Å | |||||||||
 Authors Authors | May AJ / Pothula KR / Janowska K / Acharya P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2023 Journal: J Virol / Year: 2023Title: Structures of Langya Virus Fusion Protein Ectodomain in Pre- and Postfusion Conformation. Authors: Aaron J May / Karunakar Reddy Pothula / Katarzyna Janowska / Priyamvada Acharya /  Abstract: Langya virus (LayV) is a paramyxovirus in the genus, closely related to the deadly Nipah (NiV) and Hendra (HeV) viruses, that was identified in August 2022 through disease surveillance following ...Langya virus (LayV) is a paramyxovirus in the genus, closely related to the deadly Nipah (NiV) and Hendra (HeV) viruses, that was identified in August 2022 through disease surveillance following animal exposure in eastern China. Paramyxoviruses present two glycoproteins on their surface, known as attachment and fusion proteins, that mediate entry into cells and constitute the primary antigenic targets for immune response. Here, we determine cryo-electron microscopy (cryo-EM) structures of the uncleaved LayV fusion protein (F) ectodomain in pre- and postfusion conformations. The LayV-F protein exhibits pre- and postfusion architectures that, despite being highly conserved across paramyxoviruses, show differences in their surface properties, in particular at the apex of the prefusion trimer, that may contribute to antigenic variability. While dramatic conformational changes were visualized between the pre- and postfusion forms of the LayV-F protein, several domains remained invariant, held together by highly conserved disulfides. The LayV-F fusion peptide (FP) is buried within a highly conserved, hydrophobic interprotomer pocket in the prefusion state and is notably less flexible than the rest of the protein, highlighting its "spring-loaded" state and suggesting that the mechanism of pre-to-post transition must involve perturbations to the pocket and release of the fusion peptide. Together, these results offer a structural basis for how the Langya virus fusion protein compares to its Henipavirus relatives and propose a mechanism for the initial step of pre- to postfusion conversion that may apply more broadly to paramyxoviruses. The genus is quickly expanding into new animal hosts and geographic locations. This study compares the structure and antigenicity of the Langya virus fusion protein to other henipaviruses, which have important vaccine and therapeutic development implications. Furthermore, the study proposes a new mechanism to explain the early steps of the fusion initiation process that can be more broadly applied to the family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29029.map.gz emd_29029.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29029-v30.xml emd-29029-v30.xml emd-29029.xml emd-29029.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29029_fsc.xml emd_29029_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29029.png emd_29029.png | 51.9 KB | ||
| Filedesc metadata |  emd-29029.cif.gz emd-29029.cif.gz | 5.8 KB | ||
| Others |  emd_29029_additional_1.map.gz emd_29029_additional_1.map.gz emd_29029_half_map_1.map.gz emd_29029_half_map_1.map.gz emd_29029_half_map_2.map.gz emd_29029_half_map_2.map.gz | 59.5 MB 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29029 http://ftp.pdbj.org/pub/emdb/structures/EMD-29029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29029 | HTTPS FTP |
-Validation report
| Summary document |  emd_29029_validation.pdf.gz emd_29029_validation.pdf.gz | 759.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29029_full_validation.pdf.gz emd_29029_full_validation.pdf.gz | 758.6 KB | Display | |
| Data in XML |  emd_29029_validation.xml.gz emd_29029_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_29029_validation.cif.gz emd_29029_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29029 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29029 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29029 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29029 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29029.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29029.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final 3D refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened map for model building
| File | emd_29029_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map for model building | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_29029_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_29029_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Langya Virus Fusion Protein Ectodomain Prefusion Conformation
| Entire | Name: Langya Virus Fusion Protein Ectodomain Prefusion Conformation |
|---|---|
| Components |
|
-Supramolecule #1: Langya Virus Fusion Protein Ectodomain Prefusion Conformation
| Supramolecule | Name: Langya Virus Fusion Protein Ectodomain Prefusion Conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Langya virus Langya virus |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Fusion Protein
| Macromolecule | Name: Fusion Protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Langya virus Langya virus |
| Molecular weight | Theoretical: 59.02577 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SLHYDSLSKV GIIKGLTYNY KIKGSPSTKL MVVKLIPNID GVRNCTQKQF DEYKNLVKNV LEPVKLALNA MLDNVKSGNN KYRFAGAIM AGVALGVATA ATVTAGIALH RSNENAQAIA NMKNAIQNTN EAVKQLQLAN KQTLAVIDTI RGEINNNIIP V INQLSCDT ...String: SLHYDSLSKV GIIKGLTYNY KIKGSPSTKL MVVKLIPNID GVRNCTQKQF DEYKNLVKNV LEPVKLALNA MLDNVKSGNN KYRFAGAIM AGVALGVATA ATVTAGIALH RSNENAQAIA NMKNAIQNTN EAVKQLQLAN KQTLAVIDTI RGEINNNIIP V INQLSCDT IGLSVGIKLT QYYSEILTAF GPALQNPVNT RITIQAISSV FNRNFDELLK IMGYTSGDLY EILHSGLIRG NI IDVDVEA GYIALEIEFP NLTLVPNAVV QELMPISYNV DGDEWVTLVP RFVLTRTTLL SNIDTSRCTV TESSVICDND YAL PMSYEL IGCLQGDTSK CAREKVVSSY VPRFALSDGL VYANCLNTIC RCMDTDTPIS QSLGTTVSLL DNKKCLVYQV GDIL ISVGS YLGEGEYSAD NVELGPPVVI DKIDIGNQLA GINQTLQNAE DYIEKSEEFL KGINPSIGSG YIPEAPRDGQ AYVRK DGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHSAW SHPQFEKGGG SGGGGSGGSA WSHPQFEK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6179 / Average electron dose: 56.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: SwissModel / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL |
| Output model |  PDB-8fej: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)