+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 4 | ||||||||||||
 マップデータ マップデータ | EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 4 | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | Extracellular matrix protein adhesin A Bacterial Adhesin Glycosylated protein Trimeric Autotransporter / CELL ADHESION | ||||||||||||
| 生物種 |  Aggregatibacter actinomycetemcomitans (バクテリア) Aggregatibacter actinomycetemcomitans (バクテリア) | ||||||||||||
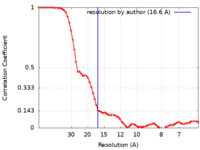
| 手法 | サブトモグラム平均法 / ネガティブ染色法 / 解像度: 16.6 Å | ||||||||||||
 データ登録者 データ登録者 | Ruiz T / Radermacher M / Mintz KP / Tang-Siegel GG | ||||||||||||
| 資金援助 |  米国, 3件 米国, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: J Bacteriol / 年: 2022 ジャーナル: J Bacteriol / 年: 2022タイトル: Serotype-Specific Sugars Impact Structure but Not Functions of the Trimeric Autotransporter Adhesin EmaA of Aggregatibacter actinomycetemcomitans. 著者: Gaoyan G Tang-Siegel / Michael Radermacher / Keith P Mintz / Teresa Ruiz /  要旨: The human oral pathobiont Aggregatibacter actinomycetemcomitans expresses multiple virulence factors, including the trimeric, extracellular matrix protein adhesin A (EmaA). The posttranslational ...The human oral pathobiont Aggregatibacter actinomycetemcomitans expresses multiple virulence factors, including the trimeric, extracellular matrix protein adhesin A (EmaA). The posttranslational modification of EmaA is proposed to be dependent on the sugars and enzymes associated with -polysaccharide (O-PS) synthesis of the lipopolysaccharide (LPS). This modification is important for the structure and function of this adhesin. To determine if the composition of the sugars alters structure and/or function, the prototypic 202-kDa protein was expressed in a non-serotype b, mutant strain. The transformed strain displayed EmaA adhesins similar in appearance to the prototypic adhesin as observed by two-dimensional (2D) electron microscopy of whole-mount negatively stained bacterial preparations. Biochemical analysis indicated that the protein monomers were posttranslationally modified. 3D electron tomographic reconstruction and structure analyses of the functional domain revealed three well-defined subdomains (SI, SII, and SIII) with a linker region between SII and SIII. Structural changes were observed in all three subdomains and the linker region of the adhesins synthesized compared with the known structure. These changes, however, did not affect the ability of the strain to bind collagen or form biofilms. The data suggest that changes in the composition of the glycan moiety alter the 3D structure of the molecule without negatively affecting the function(s) associated with this adhesin. The human oral pathogen A. actinomycetemcomitans is a causative agent of periodontal and several systemic diseases. EmaA is a trimeric autotransporter protein adhesin important for colonization by this pathobiont . This adhesin is modified with sugars associated with the -polysaccharide (O-PS), and the modification is mediated using the enzymes involved in lipopolysaccharide (LPS) biosynthesis. The interaction with collagen is not mediated by the specific binding between the glycans and collagen but is attributed to changes in the final quaternary structure necessary to maintain an active adhesin. In this study, we have determined that the composition of the sugars utilized in the posttranslational modification of this adhesin is exchangeable without compromising functional activities. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_28840.map.gz emd_28840.map.gz | 23.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-28840-v30.xml emd-28840-v30.xml emd-28840.xml emd-28840.xml | 23.5 KB 23.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_28840_fsc.xml emd_28840_fsc.xml | 8.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_28840.png emd_28840.png | 26.3 KB | ||
| Filedesc metadata |  emd-28840.cif.gz emd-28840.cif.gz | 7.7 KB | ||
| その他 |  emd_28840_half_map_1.map.gz emd_28840_half_map_1.map.gz emd_28840_half_map_2.map.gz emd_28840_half_map_2.map.gz | 24.3 MB 24.3 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28840 http://ftp.pdbj.org/pub/emdb/structures/EMD-28840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28840 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_28840_validation.pdf.gz emd_28840_validation.pdf.gz | 356.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_28840_full_validation.pdf.gz emd_28840_full_validation.pdf.gz | 355.7 KB | 表示 | |
| XML形式データ |  emd_28840_validation.xml.gz emd_28840_validation.xml.gz | 13.3 KB | 表示 | |
| CIF形式データ |  emd_28840_validation.cif.gz emd_28840_validation.cif.gz | 17.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28840 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28840 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_28840.map.gz / 形式: CCP4 / 大きさ: 25.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_28840.map.gz / 形式: CCP4 / 大きさ: 25.8 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Classification and subtomogram averaging - Class 4 | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 3.08 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: EmaA (extracellular matrix protein adhesin A) of Aggregatibacter...
| ファイル | emd_28840_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Half-Map-Odd - Class 4 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: EmaA (extracellular matrix protein adhesin A) of Aggregatibacter...
| ファイル | emd_28840_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Half-Map-Even - Class 4 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : EmaA (extracellular matrix protein adhesin A) of Aggregatibacter ...
| 全体 | 名称: EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Class 4 |
|---|---|
| 要素 |
|
-超分子 #1: EmaA (extracellular matrix protein adhesin A) of Aggregatibacter ...
| 超分子 | 名称: EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain expressed in a serotype a strain - Class 4 タイプ: organelle_or_cellular_component / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Aggregatibacter actinomycetemcomitans (バクテリア) Aggregatibacter actinomycetemcomitans (バクテリア) |
-分子 #1: EmaA (extracellular matrix protein adhesin A) of Aggregatibacter ...
| 分子 | 名称: EmaA (extracellular matrix protein adhesin A) of Aggregatibacter actinomycetemcomitans serotype b strain タイプ: other / ID: 1 / 分類: other |
|---|---|
| 由来(天然) | 生物種:  Aggregatibacter actinomycetemcomitans (バクテリア) Aggregatibacter actinomycetemcomitans (バクテリア) |
| 配列 | 文字列: MNKVFKVIWC KTSQTWIAVS ELSKAFSLST TTDIPKKTKI FIAAAPLLFL SFNTNAYIAI GSVENNSVK SEGAEASPNK RKGSQALNYY NPGSKSYDDK DKPSNPERRY SNGEAYGIAI G KNTDVRDS SKDSNGIALG DYSKATGGLA MALGSFSRAE KNGGIAIGIA ...文字列: MNKVFKVIWC KTSQTWIAVS ELSKAFSLST TTDIPKKTKI FIAAAPLLFL SFNTNAYIAI GSVENNSVK SEGAEASPNK RKGSQALNYY NPGSKSYDDK DKPSNPERRY SNGEAYGIAI G KNTDVRDS SKDSNGIALG DYSKATGGLA MALGSFSRAE KNGGIAIGIA SRSSGINSLA MM RQSAATG DYSTAIGSVA WAAGQSSFAL GASATAKGNQ SIAIGSLEQK ISPNGSGVPI TKY NGLDNT QTNGNRSMAL GTAAKTNGDD SFAIGYKAHT GEFKVEHDNY LKENVTSPDL SKKA DKAIA VGTSALAQKE SAIAFGYQAN ASGINAISLG ANAKASQDNV VAIGKDATAT ESGSM AIGQ GAKSTFKNSL ALGTGTIVNS VDGGQSKFTA QNYDANNGVV AVANAGKERR IINVAG GRN DTDAVNVAQL KFVNDNLAKS IAGAGYNGYE TDGHTYKAPV FSIKNTNYHD VKTAVEA AQ TNYVSVNSTN TAADSNYDNK GAKAVGSIAL GEKATTGRAA MNSIAIGLNS NVSGQNTV A LGANITATTN GSVILGNSST TEGSHPVSNV SSATVNGYTY SGFTGTVKES GHFVSIGSK GNERQIKNVA AGNVAANSTD AVNGSQLFAV ASRVEQGWQI TSGVENGGTQ NGAASTATIK PSNQVKLLA GKNLAVKQNG TNFTFSTQEN VTFTNVTTQD LTATGNTTVK NFSVQNGGTI N MGNNRITG VAEGTQDDDA VNFKQLKSLL GGSASTEIVE KKAAQAGDEN LADISVANGK NA GDMGAKY EVSVSKKAVQ SAAKEAVKVT GSAPINVNKT DVNGVDTYAV TFNGTEAAKS IPL TYKANG SGDKTVMLDK GLNFTNGMMT TASVANDGVM KYDVNLSTIK VEDGKAAVAG TPGT NGANG TDGKDGVATV KNVVEALNNA AWTITASKSD GEVVSNASNS VKNGDTVTYD AGKNI KITQ RDKKFSFATK DNVEFTSVTT GNTKLTGNGV EITNGPKLTQ SGVDAGGKKI TNVADG VIA ANSKDAVNGG QLFAETAKAK TTVEKGDDNI QITSETATDG HINYKVALNP SLTVGPR TN GHPITIDGNN GYITGLTNTS WTGAPTTGRA ATEDQLSIVD KKFDNKVSLG GDNGSTTE K SLSHNGGIKF NIKGGDSQKY VTTSGSGDDV TVDLAQTTKN KIDNAADKDL ANITDNGKK VITALGAVVK AADSTITVTD ETDNTTGQKT YKIKANIPTP EKTAMAPGNN TTIEGDGSAA NPFKVNLKD DLALGQKDAN GVTGKDSSIK VNGKDGSGVA INGKDGSIAL NGKDGANPVT I KTAQGPAG VNETNPKDRL MVNNDAVATL KDGLKFAGDN STEVITKTLN QKLEIVGGAD KN KLSDNNI GVNANNGKLE VKLAKELNEL TSAQFKNGDN TTVINGNGIT ITPKDPTKAV SLT DKGLNN GGNQIVNIDS GLKQADGSTV ALKDASGDTL KNAANIGDLQ KSINDITDAS KNGG FGLSD DNGATAKANL GETVKVKGDG SVITKVVTDN GKPTLQVGLS NDITVGDDAQ AGTIS VKGE NGKDGVSING KEASVTFAKD GQPGMSIAAT RSADGKDALT LKGKDGKDGI SFQEDG RIT QVADGVNDKD AVNKSQLDRS IAQAKSGVSA GKNITVTPQK NADGSTTYTV ETQKDVE FS TVKTGDTTLD SNGVNINGGP SVTKDGIHAN DKKITGVKDG EISAHSKEAV NGSQLHQT N QNVTNLANNV DKGLNFQGDN QEVTVNRKLG DQLNIRGGAD PKKLTQNNIG VTADKNGTM TVQLAKEVNL GADGSLTVGN TTVNNDGVTI KDGPSMTSHG INAGGKRIAN VAKGKAPTDA VNMSQLQDV GSAINNRIDN IDKRVKKMDK RRKAGTASAL ATAGLMQPHR DGQSALVAAV G QYQSETAV AVGYSRISDN GKYGVKVSFS TNSQGEVGGT AGAGYFW |
-実験情報
-構造解析
| 手法 | ネガティブ染色法 |
|---|---|
 解析 解析 | サブトモグラム平均法 |
| 試料の集合状態 | cell |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 構成要素:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 染色 | タイプ: NEGATIVE / 材質: NanoW 詳細: Electron microscopy grids were prepared as previously described. Briefly, a 5 ul aliquot of bacterial suspension was placed on either 300 or 200 mesh carbon-coated grids, and deep stained ...詳細: Electron microscopy grids were prepared as previously described. Briefly, a 5 ul aliquot of bacterial suspension was placed on either 300 or 200 mesh carbon-coated grids, and deep stained with NanoW (Nanoprobes, Yaphank, NY). For 3D electron tomography, the grids were pretreated with Poly-L-lysine (1000-5000 Da. Sigma, St. Louis, MO) and colloidal gold (SPI, West Chester, PA) to be used as fiducial markers. | |||||||||
| グリッド | モデル: Homemade / 材質: COPPER / メッシュ: 200 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS / 前処理 - タイプ: PLASMA CLEANING / 前処理 - 時間: 20 sec. / 前処理 - 雰囲気: OTHER |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TECNAI 12 |
|---|---|
| 撮影 | フィルム・検出器のモデル: TVIPS TEMCAM-F216 (2k x 2k) 実像数: 80 / 平均電子線量: 3.0 e/Å2 詳細: Data were collected using a Tecnai 12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 cathode (Kimball Physics, Wilton, NH), operated in point-mode, a 2048 by 2048 pixel CCD ...詳細: Data were collected using a Tecnai 12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 cathode (Kimball Physics, Wilton, NH), operated in point-mode, a 2048 by 2048 pixel CCD camera with a pixel size of 14 um, (TVIPS, Gauting, Germany) and a dual axis tilt tomography holder (Fischione, Export, PA). All images were recorded on the CCD camera at an acceleration voltage of 100 kV and a nominal magnification of 42,000, which corresponds to 0.308 nm pixel size on the specimen scale. Tomographic tilt series were acquired at least within a +/-64 degree angular range in 2 degree angular intervals. Data were collected under low dose exposure conditions (0.10 e-/nm2 for 2D imaging and 0.03 e-/nm2 per image for the tomographic tilt series data) as previously described. |
| 電子線 | 加速電圧: 100 kV / 電子線源: LAB6 |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 1.6 µm / 最小 デフォーカス(公称値): 1.3 µm / 倍率(公称値): 42000 |
| 試料ステージ | 試料ホルダーモデル: FISCHIONE INSTRUMENTS DUAL AXIS TOMOGRAPHY HOLDER ホルダー冷却材: NITROGEN |
 ムービー
ムービー コントローラー
コントローラー











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)

































 IMOD
IMOD