+ Open data
Open data
- Basic information
Basic information
| Entry | 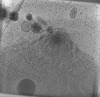 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Toxoplasma gondii apical complex (non-stimulated) | |||||||||
 Map data Map data | Toxoplasma gondii apical complex (non-stimulated) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Toxoplasma gondii / apical complex / non-stimulated / CELL INVASION | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Segev-Zarko L / Sun SY / Chiu W / Boothroyd JC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: PNAS Nexus / Year: 2022 Journal: PNAS Nexus / Year: 2022Title: Cryo-electron tomography with mixed-scale dense neural networks reveals key steps in deployment of invasion machinery. Authors: Li-Av Segev-Zarko / Peter D Dahlberg / Stella Y Sun / Daniël M Pelt / Chi Yong Kim / Elizabeth S Egan / James A Sethian / Wah Chiu / John C Boothroyd /   Abstract: Host cell invasion by intracellular, eukaryotic parasites within the phylum Apicomplexa is a remarkable and active process involving the coordinated action of apical organelles and other structures. ...Host cell invasion by intracellular, eukaryotic parasites within the phylum Apicomplexa is a remarkable and active process involving the coordinated action of apical organelles and other structures. To date, capturing how these structures interact during invasion has been difficult to observe in detail. Here, we used cryogenic electron tomography to image the apical complex of tachyzoites under conditions that mimic resting parasites and those primed to invade through stimulation with calcium ionophore. Through the application of mixed-scale dense networks for image processing, we developed a highly efficient pipeline for annotation of tomograms, enabling us to identify and extract densities of relevant subcellular organelles and accurately analyze features in 3-D. The results reveal a dramatic change in the shape of the anteriorly located apical vesicle upon its apparent fusion with a rhoptry that occurs only in the stimulated parasites. We also present information indicating that this vesicle originates from the vesicles that parallel the intraconoidal microtubules and that the latter two structures are linked by a novel tether. We show that a rosette structure previously proposed to be involved in rhoptry secretion is associated with apical vesicles beyond just the most anterior one. This result, suggesting multiple vesicles are primed to enable rhoptry secretion, may shed light on the mechanisms employs to enable repeated invasion attempts. Using the same approach, we examine merozoites and show that they too possess an apical vesicle just beneath a rosette, demonstrating evolutionary conservation of this overall subcellular organization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28140.map.gz emd_28140.map.gz | 179 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28140-v30.xml emd-28140-v30.xml emd-28140.xml emd-28140.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28140.png emd_28140.png | 135.1 KB | ||
| Filedesc metadata |  emd-28140.cif.gz emd-28140.cif.gz | 4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28140 http://ftp.pdbj.org/pub/emdb/structures/EMD-28140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28140 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28140 | HTTPS FTP |
-Validation report
| Summary document |  emd_28140_validation.pdf.gz emd_28140_validation.pdf.gz | 451.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28140_full_validation.pdf.gz emd_28140_full_validation.pdf.gz | 450.8 KB | Display | |
| Data in XML |  emd_28140_validation.xml.gz emd_28140_validation.xml.gz | 7.6 KB | Display | |
| Data in CIF |  emd_28140_validation.cif.gz emd_28140_validation.cif.gz | 10.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28140 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28140 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28140 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28140 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28140.map.gz / Format: CCP4 / Size: 355.1 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_28140.map.gz / Format: CCP4 / Size: 355.1 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Toxoplasma gondii apical complex (non-stimulated) | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 13.84 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : The apical complex of Toxoplasma gondii (non-stimulated)
| Entire | Name: The apical complex of Toxoplasma gondii (non-stimulated) |
|---|---|
| Components |
|
-Supramolecule #1: The apical complex of Toxoplasma gondii (non-stimulated)
| Supramolecule | Name: The apical complex of Toxoplasma gondii (non-stimulated) type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 / Component - Concentration: 1.0 x / Component - Name: Endo buffer Details: 45 mM potassium sulfate, 106 mM sucrose, 10 mM magnesium sulfate, 20 mM Tris buffer pH 7.2, 5 mM glucose and 0.35% bovine serum albumin |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 297 K / Instrument: LEICA EM GP |
| Details | Tachyzoites were released from heavily infected monolayers of HFFs by mechanical disruption of the monolayers using disposable scrapers and passage through a 25-gauge syringe into HPEB |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: EMS / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 26000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number images used: 61 IMOD / Number images used: 61 |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)