[English] 日本語
 Yorodumi
Yorodumi- EMDB-27404: CryoEM structure of the 1:1 ADP-tetrafluoroaluminate stabilized n... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the 1:1 ADP-tetrafluoroaluminate stabilized nitrogenase complex from Azotobacter vinelandii | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Nitrogenase / metalloenzyme / metal binding protein / oxidoreductase | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / 4 iron, 4 sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.48 Å | ||||||||||||
 Authors Authors | Warmack RA / Rees DC | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Protoc / Year: 2024 Journal: Nat Protoc / Year: 2024Title: Anaerobic cryoEM protocols for air-sensitive nitrogenase proteins. Authors: Rebeccah A Warmack / Belinda B Wenke / Thomas Spatzal / Douglas C Rees /  Abstract: Single-particle cryo-electron microscopy (cryoEM) provides an attractive avenue for advancing our atomic resolution understanding of materials, molecules and living systems. However, the vast ...Single-particle cryo-electron microscopy (cryoEM) provides an attractive avenue for advancing our atomic resolution understanding of materials, molecules and living systems. However, the vast majority of published cryoEM methodologies focus on the characterization of aerobically purified samples. Air-sensitive enzymes and microorganisms represent important yet understudied systems in structural biology. We have recently demonstrated the success of an anaerobic single-particle cryoEM workflow applied to the air-sensitive nitrogenase enzymes. In this protocol, we detail the use of Schlenk lines and anaerobic chambers to prepare samples, including a protein tag for monitoring sample exposure to oxygen in air. We describe how to use a plunge freezing apparatus inside of a soft-sided vinyl chamber of the type we routinely use for anaerobic biochemistry and crystallography of oxygen-sensitive proteins. Manual control of the airlock allows for introduction of liquid cryogens into the tent. A custom vacuum port provides slow, continuous evacuation of the tent atmosphere to avoid accumulation of flammable vapors within the enclosed chamber. These methods allowed us to obtain high-resolution structures of both nitrogenase proteins using single-particle cryoEM. The procedures involved can be generally subdivided into a 4 d anaerobic sample generation procedure, and a 1 d anaerobic cryoEM sample preparation step, followed by conventional cryoEM imaging and processing steps. As nitrogen is a substrate for nitrogenase, the Schlenk lines and anaerobic chambers described in this procedure are operated under an argon atmosphere; however, the system and these procedures are compatible with other controlled gas environments. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27404.map.gz emd_27404.map.gz | 374.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27404-v30.xml emd-27404-v30.xml emd-27404.xml emd-27404.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27404_fsc.xml emd_27404_fsc.xml | 18.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_27404.png emd_27404.png | 63.8 KB | ||
| Filedesc metadata |  emd-27404.cif.gz emd-27404.cif.gz | 7.1 KB | ||
| Others |  emd_27404_half_map_1.map.gz emd_27404_half_map_1.map.gz emd_27404_half_map_2.map.gz emd_27404_half_map_2.map.gz | 677.3 MB 677.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27404 http://ftp.pdbj.org/pub/emdb/structures/EMD-27404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27404 | HTTPS FTP |
-Validation report
| Summary document |  emd_27404_validation.pdf.gz emd_27404_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27404_full_validation.pdf.gz emd_27404_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_27404_validation.xml.gz emd_27404_validation.xml.gz | 28.3 KB | Display | |
| Data in CIF |  emd_27404_validation.cif.gz emd_27404_validation.cif.gz | 37.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27404 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27404 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27404 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27404 | HTTPS FTP |
-Related structure data
| Related structure data |  8dfcMC  8dbyC  8dfdC  8tc3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27404.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27404.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27404_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
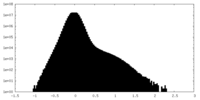
| Density Histograms |
-Half map: #2
| File | emd_27404_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Heterohexameric nitrogenase MoFe-protein - Fe-protein 1:1 complex...
+Supramolecule #1: Heterohexameric nitrogenase MoFe-protein - Fe-protein 1:1 complex...
+Macromolecule #1: Nitrogenase molybdenum-iron protein alpha chain
+Macromolecule #2: Nitrogenase molybdenum-iron protein beta chain
+Macromolecule #3: Nitrogenase iron protein 1
+Macromolecule #4: iron-sulfur-molybdenum cluster with interstitial carbon
+Macromolecule #5: 3-HYDROXY-3-CARBOXY-ADIPIC ACID
+Macromolecule #6: FE(8)-S(7) CLUSTER
+Macromolecule #7: FE (III) ION
+Macromolecule #8: IRON/SULFUR CLUSTER
+Macromolecule #9: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #10: TETRAFLUOROALUMINATE ION
+Macromolecule #11: MAGNESIUM ION
+Macromolecule #12: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: -3.0 µm / Nominal defocus min: -0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)