[English] 日本語
 Yorodumi
Yorodumi- EMDB-2717: Electron cryo-microscopy of T7 bacteriophage tail after DNA ejection -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2717 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
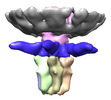
| Title | Electron cryo-microscopy of T7 bacteriophage tail after DNA ejection | |||||||||
 Map data Map data | Reconstruction of T7 tail after DNA ejection | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / virus infection / DNA ejection / tail | |||||||||
| Biological species |   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Gonzalez-Garcia VA / Pulido-Cid M / Garcia-Doval C / van Raaij MJ / Martin-Benito J / Cuervo A / Carrascosa JL | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2015 Journal: J Biol Chem / Year: 2015Title: Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. Authors: Verónica A González-García / Mar Pulido-Cid / Carmela Garcia-Doval / Rebeca Bocanegra / Mark J van Raaij / Jaime Martín-Benito / Ana Cuervo / José L Carrascosa /  Abstract: The majority of bacteriophages protect their genetic material by packaging the nucleic acid in concentric layers to an almost crystalline concentration inside protein shells (capsid). This highly ...The majority of bacteriophages protect their genetic material by packaging the nucleic acid in concentric layers to an almost crystalline concentration inside protein shells (capsid). This highly condensed genome also has to be efficiently injected into the host bacterium in a process named ejection. Most phages use a specialized complex (often a tail) to deliver the genome without disrupting cell integrity. Bacteriophage T7 belongs to the Podoviridae family and has a short, non-contractile tail formed by a tubular structure surrounded by fibers. Here we characterize the kinetics and structure of bacteriophage T7 DNA delivery process. We show that T7 recognizes lipopolysaccharides (LPS) from Escherichia coli rough strains through the fibers. Rough LPS acts as the main phage receptor and drives DNA ejection in vitro. The structural characterization of the phage tail after ejection using cryo-electron microscopy (cryo-EM) and single particle reconstruction methods revealed the major conformational changes needed for DNA delivery at low resolution. Interaction with the receptor causes fiber tilting and opening of the internal tail channel by untwisting the nozzle domain, allowing release of DNA and probably of the internal head proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2717.map.gz emd_2717.map.gz | 784.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2717-v30.xml emd-2717-v30.xml emd-2717.xml emd-2717.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2717.png emd_2717.png | 89.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2717 http://ftp.pdbj.org/pub/emdb/structures/EMD-2717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2717 | HTTPS FTP |
-Validation report
| Summary document |  emd_2717_validation.pdf.gz emd_2717_validation.pdf.gz | 236.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2717_full_validation.pdf.gz emd_2717_full_validation.pdf.gz | 235.5 KB | Display | |
| Data in XML |  emd_2717_validation.xml.gz emd_2717_validation.xml.gz | 4.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2717 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2717 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2717 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2717 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2717.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2717.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of T7 tail after DNA ejection | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
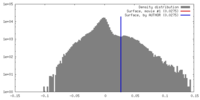
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : T7 tail after DNA ejection
| Entire | Name: T7 tail after DNA ejection |
|---|---|
| Components |
|
-Supramolecule #1000: T7 tail after DNA ejection
| Supramolecule | Name: T7 tail after DNA ejection / type: sample / ID: 1000 Oligomeric state: Oligomer build of 5 components: gp8 (12mer), gp11 (12mer), gp12 (6mer), gp17 (3mer), gp10 (n/a) Number unique components: 5 |
|---|
-Macromolecule #1: gp8
| Macromolecule | Name: gp8 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) |
-Macromolecule #2: gp11
| Macromolecule | Name: gp11 / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) |
-Macromolecule #3: gp12
| Macromolecule | Name: gp12 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) |
-Macromolecule #4: gp17
| Macromolecule | Name: gp17 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) |
-Macromolecule #5: gp10
| Macromolecule | Name: gp10 / type: protein_or_peptide / ID: 5 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.8 Details: 50 mM TrisHCl, 10 mM MgCl2, 100 mM NaCl, 0.25mg/ml LPS from E. coli EH 100 Ra mutant |
| Grid | Details: R2/2 Quantifoil coated with a thin carbon layer |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC Method: Samples were applied to grids for 1 minute, blotted, and plunged into liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 93 K |
| Date | Oct 11, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 2 µm / Number real images: 463 / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 108696 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were manually selected |
|---|---|
| CTF correction | Details: Each micrograph |
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Software - Name: XMIPP, EMAN / Details: Final map was calculated from 3 averaged data sets / Number images used: 1781 |
| Final two d classification | Number classes: 4 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)