[English] 日本語
 Yorodumi
Yorodumi- EMDB-27121: CryoEM structure of human orphan GPCR GPR179 in complex with extr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human orphan GPCR GPR179 in complex with extracellular matrix protein pikachurin | |||||||||
 Map data Map data | Structure of an orphan GPCR complexed with ECM protein | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR complex with ECM / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinterstitial matrix / photoreceptor ribbon synapse / glycosaminoglycan binding / positive regulation of cell-substrate adhesion / presynaptic active zone / basement membrane / synaptic cleft / visual perception / extracellular matrix organization / cell projection ...interstitial matrix / photoreceptor ribbon synapse / glycosaminoglycan binding / positive regulation of cell-substrate adhesion / presynaptic active zone / basement membrane / synaptic cleft / visual perception / extracellular matrix organization / cell projection / G protein-coupled receptor activity / postsynaptic membrane / dendrite / calcium ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Patil DN / Martemyanov KA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Signal / Year: 2023 Journal: Sci Signal / Year: 2023Title: Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179. Authors: Dipak N Patil / Serena Pantalone / Yan Cao / Thibaut Laboute / Scott J Novick / Shikha Singh / Simone Savino / Silvia Faravelli / Francesca Magnani / Patrick R Griffin / Appu K Singh / ...Authors: Dipak N Patil / Serena Pantalone / Yan Cao / Thibaut Laboute / Scott J Novick / Shikha Singh / Simone Savino / Silvia Faravelli / Francesca Magnani / Patrick R Griffin / Appu K Singh / Federico Forneris / Kirill A Martemyanov /    Abstract: Precise synapse formation is essential for normal functioning of the nervous system. Retinal photoreceptors establish selective contacts with bipolar cells, aligning the neurotransmitter release ...Precise synapse formation is essential for normal functioning of the nervous system. Retinal photoreceptors establish selective contacts with bipolar cells, aligning the neurotransmitter release apparatus with postsynaptic signaling cascades. This involves transsynaptic assembly between the dystroglycan-dystrophin complex on the photoreceptor and the orphan receptor GPR179 on the bipolar cell, which is mediated by the extracellular matrix protein pikachurin (also known as EGFLAM). This complex plays a critical role in the synaptic organization of photoreceptors and signal transmission, and mutations affecting its components cause blinding disorders in humans. Here, we investigated the structural organization and molecular mechanisms by which pikachurin orchestrates transsynaptic assembly and solved structures of the human pikachurin domains by x-ray crystallography and of the GPR179-pikachurin complex by single-particle, cryo-electron microscopy. The structures reveal molecular recognition principles of pikachurin by the Cache domains of GPR179 and show how the interaction is involved in the transsynaptic alignment of the signaling machinery. Together, these data provide a structural basis for understanding the synaptic organization of photoreceptors and ocular pathology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27121.map.gz emd_27121.map.gz | 398.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27121-v30.xml emd-27121-v30.xml emd-27121.xml emd-27121.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27121.png emd_27121.png | 43.5 KB | ||
| Filedesc metadata |  emd-27121.cif.gz emd-27121.cif.gz | 5.9 KB | ||
| Others |  emd_27121_half_map_1.map.gz emd_27121_half_map_1.map.gz emd_27121_half_map_2.map.gz emd_27121_half_map_2.map.gz | 391.4 MB 391.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27121 http://ftp.pdbj.org/pub/emdb/structures/EMD-27121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27121 | HTTPS FTP |
-Validation report
| Summary document |  emd_27121_validation.pdf.gz emd_27121_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27121_full_validation.pdf.gz emd_27121_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_27121_validation.xml.gz emd_27121_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_27121_validation.cif.gz emd_27121_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27121 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27121 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27121 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27121 | HTTPS FTP |
-Related structure data
| Related structure data |  8d1bMC  7zc9C  7zcbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27121.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27121.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of an orphan GPCR complexed with ECM protein | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||
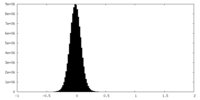
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_27121_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
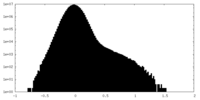
| Density Histograms |
-Half map: Half Map 2
| File | emd_27121_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
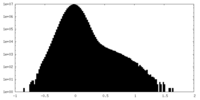
| Density Histograms |
- Sample components
Sample components
-Entire : complex of GPR179 with pikachurin
| Entire | Name: complex of GPR179 with pikachurin |
|---|---|
| Components |
|
-Supramolecule #1: complex of GPR179 with pikachurin
| Supramolecule | Name: complex of GPR179 with pikachurin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Pikachurin
| Macromolecule | Name: Pikachurin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.64225 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AAHPCVRAPC AHGGSCRPRK EGYDCDCPLG FEGLHCQKEC GNYCLNTIIE AIEIPQFIGR SYLTYDNPDI LKRVSGSRSN VFMRFKTTA KDGLLLWRGD SPMRPNSDFI SLGLRDGALV FSYNLGSGVA SIMVNGSFND GRWHRVKAVR DGQSGKITVD D YGARTGKS ...String: AAHPCVRAPC AHGGSCRPRK EGYDCDCPLG FEGLHCQKEC GNYCLNTIIE AIEIPQFIGR SYLTYDNPDI LKRVSGSRSN VFMRFKTTA KDGLLLWRGD SPMRPNSDFI SLGLRDGALV FSYNLGSGVA SIMVNGSFND GRWHRVKAVR DGQSGKITVD D YGARTGKS PGMMRQLNIN GALYVGGMKE IALHTNRQYM RGLVGCISHF TLSTDYHISL VEDAVDGKNI NTCGAC UniProtKB: Pikachurin |
-Macromolecule #2: Probable G-protein coupled receptor 179
| Macromolecule | Name: Probable G-protein coupled receptor 179 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 81.790227 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGTRGAVMPP PMWGLLGCCF VCAWALGGPR PIRSLPPLSS QVKPGSVPMQ VPLEGAEAAL AYLYSGDAQQ LSQVNCSERY EARGAGAMP GLPPSLQGAA GTLAQAANFL NMLLQANDIR ESSVEEDVEW YQALVRSVAE GDPRVYRALL TFNPPPGASH L QLALQATR ...String: MGTRGAVMPP PMWGLLGCCF VCAWALGGPR PIRSLPPLSS QVKPGSVPMQ VPLEGAEAAL AYLYSGDAQQ LSQVNCSERY EARGAGAMP GLPPSLQGAA GTLAQAANFL NMLLQANDIR ESSVEEDVEW YQALVRSVAE GDPRVYRALL TFNPPPGASH L QLALQATR TGEETILQDL SGNWVQEENP PGDLDTPALK KRVLTNDLGS LGSPKWPQAD GYVGDTQQVR LSPPFLECQE GR LRPGWLI TLSATFYGLK PDLSPEVRGQ VQMDVDLQSV DINQCASGPG WYSNTHLCDL NSTQCVPLES QGFVLGRYLC RCR PGFYGA SPSGGLEESD FQTTGQFGFP EGRSGRLLQC LPCPEGCTSC MDATPCLVEE AAVLRAAVLA CQACCMLAIF LSML VSYRC RRNKRIWASG VVLLETVLFG FLLLYFPVFI LYFKPSVFRC IALRWVRLLG FAIVYGTIIL KLYRVLQLFL SRTAQ RSAL LSSGRLLRRL GLLLLPVLGF LAVWTVGALE RGIQHAPLVI RGHTPSGRHF YLCHHDRWDY IMVVAELLLL CWGSFL CYA TRAVLSAFHE PRYMGIALHN ELLLSAAFHT ARFVLVPSLH PDWTLLLFFF HTHSTVTTTL ALIFIPKFWK LGAPPRE EM VDEVCEDELD LQHSGSYLGS SIASAWSEHS LDPGDIRDEL KKLYAQLEVH KTKEMAANNP HLPKKRGSSC QGLGRSFM R YLAEFPEALA RQHSRDSGS UniProtKB: Probable G-protein coupled receptor 179 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: alphafold |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.57 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 217570 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)