+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-2707 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | The structure of the immature M-PMV capsid in intact virus particles | |||||||||
 マップデータ マップデータ | Subtomogram averaging reconstruction of the immature Mason-Pfizer monkey virus capsid from intact virus particles | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | immature M-PMV / Mason-Pfizer Monkey virus / Retrovirus / Maturation / capsid / Gag | |||||||||
| 生物種 |  Mason-Pfizer monkey virus (ウイルス) Mason-Pfizer monkey virus (ウイルス) | |||||||||
| 手法 | サブトモグラム平均法 / クライオ電子顕微鏡法 / 解像度: 9.7 Å | |||||||||
 データ登録者 データ登録者 | Schur FKM / Hagen WJH / Rumlova M / Ruml T / Mueller B / Kraeusslich H-G / Briggs JAG | |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2015 ジャーナル: Nature / 年: 2015タイトル: Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. 著者: Florian K M Schur / Wim J H Hagen / Michaela Rumlová / Tomáš Ruml / Barbara Müller / Hans-Georg Kräusslich / John A G Briggs /   要旨: Human immunodeficiency virus type 1 (HIV-1) assembly proceeds in two stages. First, the 55 kilodalton viral Gag polyprotein assembles into a hexameric protein lattice at the plasma membrane of the ...Human immunodeficiency virus type 1 (HIV-1) assembly proceeds in two stages. First, the 55 kilodalton viral Gag polyprotein assembles into a hexameric protein lattice at the plasma membrane of the infected cell, inducing budding and release of an immature particle. Second, Gag is cleaved by the viral protease, leading to internal rearrangement of the virus into the mature, infectious form. Immature and mature HIV-1 particles are heterogeneous in size and morphology, preventing high-resolution analysis of their protein arrangement in situ by conventional structural biology methods. Here we apply cryo-electron tomography and sub-tomogram averaging methods to resolve the structure of the capsid lattice within intact immature HIV-1 particles at subnanometre resolution, allowing unambiguous positioning of all α-helices. The resulting model reveals tertiary and quaternary structural interactions that mediate HIV-1 assembly. Strikingly, these interactions differ from those predicted by the current model based on in vitro-assembled arrays of Gag-derived proteins from Mason-Pfizer monkey virus. To validate this difference, we solve the structure of the capsid lattice within intact immature Mason-Pfizer monkey virus particles. Comparison with the immature HIV-1 structure reveals that retroviral capsid proteins, while having conserved tertiary structures, adopt different quaternary arrangements during virus assembly. The approach demonstrated here should be applicable to determine structures of other proteins at subnanometre resolution within heterogeneous environments. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_2707.map.gz emd_2707.map.gz | 9.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-2707-v30.xml emd-2707-v30.xml emd-2707.xml emd-2707.xml | 9.6 KB 9.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_2707.tif emd_2707.tif | 759.3 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2707 http://ftp.pdbj.org/pub/emdb/structures/EMD-2707 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2707 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2707 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_2707_validation.pdf.gz emd_2707_validation.pdf.gz | 228 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_2707_full_validation.pdf.gz emd_2707_full_validation.pdf.gz | 227.2 KB | 表示 | |
| XML形式データ |  emd_2707_validation.xml.gz emd_2707_validation.xml.gz | 5.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2707 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2707 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2707 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2707 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_2707.map.gz / 形式: CCP4 / 大きさ: 10.2 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_2707.map.gz / 形式: CCP4 / 大きさ: 10.2 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Subtomogram averaging reconstruction of the immature Mason-Pfizer monkey virus capsid from intact virus particles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.025 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
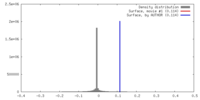
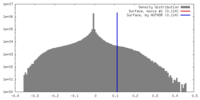
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : intact Mason-Pfizer Monkey virus particles
| 全体 | 名称: intact Mason-Pfizer Monkey virus particles |
|---|---|
| 要素 |
|
-超分子 #1000: intact Mason-Pfizer Monkey virus particles
| 超分子 | 名称: intact Mason-Pfizer Monkey virus particles / タイプ: sample / ID: 1000 詳細: The virus contained an inactivating mutation in the protease (D26N) 集合状態: Homohexameric / Number unique components: 1 |
|---|
-超分子 #1: Mason-Pfizer monkey virus
| 超分子 | 名称: Mason-Pfizer monkey virus / タイプ: virus / ID: 1 / Name.synonym: M-PMV / NCBI-ID: 11855 / 生物種: Mason-Pfizer monkey virus / ウイルスタイプ: VIRION / ウイルス・単離状態: SPECIES / ウイルス・エンベロープ: Yes / ウイルス・中空状態: No / Syn species name: M-PMV |
|---|---|
| 宿主 | 生物種:  Cercopithecidae (オナガザル) / 別称: VERTEBRATES Cercopithecidae (オナガザル) / 別称: VERTEBRATES |
| Host system | 組換細胞: HEK293T / 組換プラスミド: pSARM |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | サブトモグラム平均法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | 詳細: PBS |
|---|---|
| グリッド | 詳細: C-Flat 2/2-3C glow discharged |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / 装置: FEI VITROBOT MARK II 手法: Degassed C-Flat 2/2-3C grids were glow discharged for 30 seconds at 20 mA. Virus solution was diluted in PBS containing 10nm colloidal gold. 2 ul of this mixture was applied to a grid. Blotting time: 2 seconds |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| アライメント法 | Legacy - 非点収差: Objective lens astigmatism was corrected at nominal working magnification |
| 特殊光学系 | エネルギーフィルター - 名称: GATAN GIF 2002 |
| 日付 | 2014年2月3日 |
| 撮影 | カテゴリ: CCD / フィルム・検出器のモデル: GATAN MULTISCAN / 平均電子線量: 40 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 4.5 µm / 最小 デフォーカス(公称値): 1.5 µm / 倍率(公称値): 42000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER Tilt series - Axis1 - Min angle: -45 ° / Tilt series - Axis1 - Max angle: 60 ° |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 詳細 | Subtomogram averaging calculations were performed using the AV3 and TOM packages. Subtomograms were extracted from the surface of the virus. |
|---|---|
| 最終 再構成 | 想定した対称性 - 点群: C6 (6回回転対称) / アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 9.7 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: TOM, AV3 詳細: Reconstruction carried out using subtomogram averaging. Subtomogram averaging was performed using scripts from the TOM (Nickell et al, 2005) and AV3 (Foerster et al, 2005) packages. 使用したサブトモグラム数: 12920 |
| CTF補正 | 詳細: Phase flipping of individual tilts |
 ムービー
ムービー コントローラー
コントローラー












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)