[English] 日本語
 Yorodumi
Yorodumi- EMDB-2702: Cryo-EM structure of the mammalian 60S ribosomal subunit in compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2702 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the mammalian 60S ribosomal subunit in complex with eIF6 and Listerin | |||||||||
 Map data Map data | 60S ribosomal subunit in complex with eIF6 and Listerin E3 ligase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / ubiquitination / quality control | |||||||||
| Biological species |    Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Shao S / Hegde RS | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2014 Journal: Mol Cell / Year: 2014Title: Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Authors: Sichen Shao / Ramanujan S Hegde /  Abstract: Ribosomes stalled on aberrant mRNAs engage quality control mechanisms that degrade the partially translated nascent polypeptide. Ubiquitination of the nascent protein is mediated by the E3 ligase ...Ribosomes stalled on aberrant mRNAs engage quality control mechanisms that degrade the partially translated nascent polypeptide. Ubiquitination of the nascent protein is mediated by the E3 ligase Listerin via a mechanism involving ribosome subunit dissociation. Here, we reconstitute ribosome-associated ubiquitination with purified factors to define the minimal components and essential steps in this process. We find that the primary role of the ribosome splitting factors Hbs1, Pelota, and ABCE1 is to permit Listerin access to the nascent chain. Listerin alone can discriminate 60S- from 80S-nascent chain complexes to selectively ubiquitinate the former. Splitting factors can be bypassed by artificially removing the 40S subunit, suggesting that mere steric hindrance impedes Listerin recruitment. This was illustrated by a cryo-EM reconstruction of the 60S-Listerin complex that identifies a binding interface that clashes with the 40S ribosomal subunit. These results reveal the mechanistic logic of the core steps in a ribosome-associated quality control pathway. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2702.map.gz emd_2702.map.gz | 20.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2702-v30.xml emd-2702-v30.xml emd-2702.xml emd-2702.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2702.tif emd_2702.tif | 758.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2702 http://ftp.pdbj.org/pub/emdb/structures/EMD-2702 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2702 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2702 | HTTPS FTP |
-Validation report
| Summary document |  emd_2702_validation.pdf.gz emd_2702_validation.pdf.gz | 246.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2702_full_validation.pdf.gz emd_2702_full_validation.pdf.gz | 245.9 KB | Display | |
| Data in XML |  emd_2702_validation.xml.gz emd_2702_validation.xml.gz | 6.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2702 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2702 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2702 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2702 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2702.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2702.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 60S ribosomal subunit in complex with eIF6 and Listerin E3 ligase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
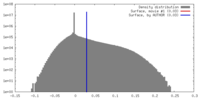
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : mammalian 60S ribosomal subunit in complex with eIF6 and Listerin...
| Entire | Name: mammalian 60S ribosomal subunit in complex with eIF6 and Listerin E3 ligase |
|---|---|
| Components |
|
-Supramolecule #1000: mammalian 60S ribosomal subunit in complex with eIF6 and Listerin...
| Supramolecule | Name: mammalian 60S ribosomal subunit in complex with eIF6 and Listerin E3 ligase type: sample / ID: 1000 / Number unique components: 3 |
|---|
-Supramolecule #1: 60S ribosomal subunit
| Supramolecule | Name: 60S ribosomal subunit / type: complex / ID: 1 / Recombinant expression: No Ribosome-details: ribosome-eukaryote: LSU 60S, LSU RNA 28S, LSU RNA 5.8S, LSU RNA 5S |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.9 MDa |
-Macromolecule #1: eukaryotic initiation factor 6
| Macromolecule | Name: eukaryotic initiation factor 6 / type: protein_or_peptide / ID: 1 / Name.synonym: eIF6 / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: Listerin
| Macromolecule | Name: Listerin / type: protein_or_peptide / ID: 2 / Name.synonym: RNF160, ZNF294 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Theoretical: 200 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pcDNA3.1 Homo sapiens (human) / Recombinant cell: HEK293T / Recombinant plasmid: pcDNA3.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 50 mM Hepes, 100 mM KAc, 5 mM MgAc2, 1 mM DTT |
|---|---|
| Grid | Details: Quantifoil R2/2 on 400 mesh Cu grid with thin carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK I / Method: Blot 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 59,000 times magnification |
| Date | Apr 1, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 1568 / Average electron dose: 25 e/Å2 Details: An in-house system was used to intercept the videos from the detector at a rate of 17 frames for 1s exposures |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 104478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | RELION movement correction processing |
|---|---|
| CTF correction | Details: each particle |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.9 Å / Resolution method: OTHER / Software - Name: CTFFIND3, RELION / Number images used: 9148 |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)