[English] 日本語
 Yorodumi
Yorodumi- EMDB-26791: Cryo-EM subtomogram average of IFT-A in anterograde IFT trains at... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM subtomogram average of IFT-A in anterograde IFT trains at 23 Angstrom resolution (Chlamydomonas reinhardtii) | |||||||||
 Map data Map data | Primary masked map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 23.0 Å | |||||||||
 Authors Authors | Jordan MA / Pigino G | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Integrative modeling reveals the molecular architecture of the Intraflagellar Transport A (IFT-A) complex Authors: McCafferty CL / Papoulas O / Jordan MA / Hoogerbrugge G / Nichols C / Pigino G / Taylor DW / Wallingford JB / Marcotte EM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26791.map.gz emd_26791.map.gz | 109.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26791-v30.xml emd-26791-v30.xml emd-26791.xml emd-26791.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26791.png emd_26791.png | 66.4 KB | ||
| Others |  emd_26791_additional_1.map.gz emd_26791_additional_1.map.gz emd_26791_additional_2.map.gz emd_26791_additional_2.map.gz | 844.3 KB 93.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26791 http://ftp.pdbj.org/pub/emdb/structures/EMD-26791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26791 | HTTPS FTP |
-Validation report
| Summary document |  emd_26791_validation.pdf.gz emd_26791_validation.pdf.gz | 300.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26791_full_validation.pdf.gz emd_26791_full_validation.pdf.gz | 299.7 KB | Display | |
| Data in XML |  emd_26791_validation.xml.gz emd_26791_validation.xml.gz | 5.2 KB | Display | |
| Data in CIF |  emd_26791_validation.cif.gz emd_26791_validation.cif.gz | 5.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26791 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26791 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26791 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26791 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26791.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26791.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary masked map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.066 Å | ||||||||||||||||||||||||||||||||||||
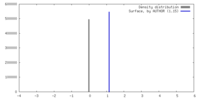
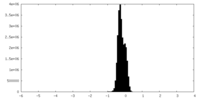
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Composite polymer map
| File | emd_26791_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite polymer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
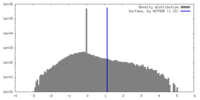
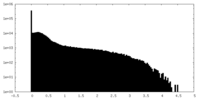
| Density Histograms |
-Additional map: Large 300px raw map
| File | emd_26791_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Large 300px raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
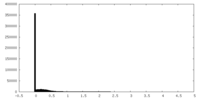
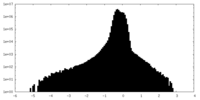
| Density Histograms |
- Sample components
Sample components
-Entire : IFT-A complex comprised of IFT43, IFT121, IFT122, IFT139, IFT140,...
| Entire | Name: IFT-A complex comprised of IFT43, IFT121, IFT122, IFT139, IFT140, and IFT144 |
|---|---|
| Components |
|
-Supramolecule #1: IFT-A complex comprised of IFT43, IFT121, IFT122, IFT139, IFT140,...
| Supramolecule | Name: IFT-A complex comprised of IFT43, IFT121, IFT122, IFT139, IFT140, and IFT144 type: complex / Chimera: Yes / ID: 1 / Parent: 0 Details: Cryo-ET subtomogram averaging of an IFT-A complex, as observed in situ in the context of an anterograde intraflagellar transport (IFT) train in intact Chlamydomonas flagella |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 291.15 K / Instrument: LEICA EM GP |
| Details | Fresh liquid cell cultures of 300 mL were grown for three to four days in TAP (Tris-acetate-phosphate) medium at 22 degrees C under a light-dark cycle with constant aeration. 3 microL undiluted Chlamydomonas cells were applied to the grid and mixed with 1 microL 10 nm colloidal gold particles (BBI solutions) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV Details: Images were recorded with an energy filter (GIF, Gatan image filter). Digital Micrograph software (Gatan) was used to tune the GIF, and SerialEM software was employed for the automated ...Details: Images were recorded with an energy filter (GIF, Gatan image filter). Digital Micrograph software (Gatan) was used to tune the GIF, and SerialEM software was employed for the automated acquisition of tomographic tilt series in low-dose mode. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-10 / Number grids imaged: 34 / Average exposure time: 2.0 sec. / Average electron dose: 1.85 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 3.0 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PEET / Number subtomograms used: 9350 |
|---|---|
| Extraction | Number tomograms: 96 / Number images used: 9350 / Method: manual picking / Software - Name:  IMOD IMODDetails: bin6 tomograms were filtered with NAD (non-linear anisotropic diffusion) in IMOD to increase contrast and facilitate particle picking |
| CTF correction | Software - Name:  IMOD IMODDetails: CTF curves were estimated with CTFPLOTTER and the data were corrected by phase-flipping with CTFPHASEFLIP, both implemented in IMOD. |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: PEET |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)