[English] 日本語
 Yorodumi
Yorodumi- EMDB-26736: Hantavirus MAPV Gn(H)/Gc protein in complex with 2 Fabs SNV-24 an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
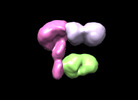
| Title | Hantavirus MAPV Gn(H)/Gc protein in complex with 2 Fabs SNV-24 and SNV-53 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Maporal virus / Maporal virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 22.0 Å | |||||||||
 Authors Authors | Binshtein E / Crowe JE | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Antigenic mapping and functional characterization of human New World hantavirus neutralizing antibodies. Authors: Taylor B Engdahl / Elad Binshtein / Rebecca L Brocato / Natalia A Kuzmina / Lucia M Principe / Steven A Kwilas / Robert K Kim / Nathaniel S Chapman / Monique S Porter / Pablo Guardado-Calvo ...Authors: Taylor B Engdahl / Elad Binshtein / Rebecca L Brocato / Natalia A Kuzmina / Lucia M Principe / Steven A Kwilas / Robert K Kim / Nathaniel S Chapman / Monique S Porter / Pablo Guardado-Calvo / Félix A Rey / Laura S Handal / Summer M Diaz / Irene A Zagol-Ikapitte / Minh H Tran / W Hayes McDonald / Jens Meiler / Joseph X Reidy / Andrew Trivette / Alexander Bukreyev / Jay W Hooper / James E Crowe /   Abstract: Hantaviruses are high-priority emerging pathogens carried by rodents and transmitted to humans by aerosolized excreta or, in rare cases, person-to-person contact. While infections in humans are ...Hantaviruses are high-priority emerging pathogens carried by rodents and transmitted to humans by aerosolized excreta or, in rare cases, person-to-person contact. While infections in humans are relatively rare, mortality rates range from 1 to 40% depending on the hantavirus species. There are currently no FDA-approved vaccines or therapeutics for hantaviruses, and the only treatment for infection is supportive care for respiratory or kidney failure. Additionally, the human humoral immune response to hantavirus infection is incompletely understood, especially the location of major antigenic sites on the viral glycoproteins and conserved neutralizing epitopes. Here, we report antigenic mapping and functional characterization for four neutralizing hantavirus antibodies. The broadly neutralizing antibody SNV-53 targets an interface between Gn/Gc, neutralizes through fusion inhibition and cross-protects against the Old World hantavirus species Hantaan virus when administered pre- or post-exposure. Another broad antibody, SNV-24, also neutralizes through fusion inhibition but targets domain I of Gc and demonstrates weak neutralizing activity to authentic hantaviruses. ANDV-specific, neutralizing antibodies (ANDV-5 and ANDV-34) neutralize through attachment blocking and protect against hantavirus cardiopulmonary syndrome (HCPS) in animals but target two different antigenic faces on the head domain of Gn. Determining the antigenic sites for neutralizing antibodies will contribute to further therapeutic development for hantavirus-related diseases and inform the design of new broadly protective hantavirus vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26736.map.gz emd_26736.map.gz | 1.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26736-v30.xml emd-26736-v30.xml emd-26736.xml emd-26736.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26736.png emd_26736.png | 33.2 KB | ||
| Others |  emd_26736_half_map_1.map.gz emd_26736_half_map_1.map.gz emd_26736_half_map_2.map.gz emd_26736_half_map_2.map.gz | 3.1 MB 3.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26736 http://ftp.pdbj.org/pub/emdb/structures/EMD-26736 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26736 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26736 | HTTPS FTP |
-Validation report
| Summary document |  emd_26736_validation.pdf.gz emd_26736_validation.pdf.gz | 564.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26736_full_validation.pdf.gz emd_26736_full_validation.pdf.gz | 564.1 KB | Display | |
| Data in XML |  emd_26736_validation.xml.gz emd_26736_validation.xml.gz | 7.9 KB | Display | |
| Data in CIF |  emd_26736_validation.cif.gz emd_26736_validation.cif.gz | 9.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26736 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26736 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26736 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26736 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26736.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26736.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.54 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_26736_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
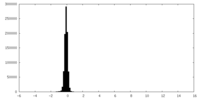
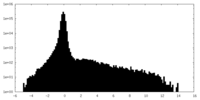
| Density Histograms |
-Half map: #2
| File | emd_26736_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
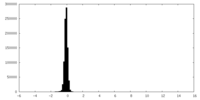
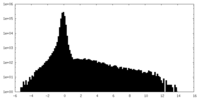
| Density Histograms |
- Sample components
Sample components
-Entire : Hanta MAPV virus Gn(H)/Gc protein in complex with 2 Fabs
| Entire | Name: Hanta MAPV virus Gn(H)/Gc protein in complex with 2 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Hanta MAPV virus Gn(H)/Gc protein in complex with 2 Fabs
| Supramolecule | Name: Hanta MAPV virus Gn(H)/Gc protein in complex with 2 Fabs type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 200 KDa |
-Supramolecule #2: Hanta MAPV virus Gn(H)/Gc protein
| Supramolecule | Name: Hanta MAPV virus Gn(H)/Gc protein / type: complex / ID: 2 / Chimera: Yes / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Maporal virus Maporal virus |
| Molecular weight | Theoretical: 150 KDa |
-Supramolecule #3: Fab
| Supramolecule | Name: Fab / type: complex / ID: 3 / Chimera: Yes / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Staining | Type: NEGATIVE / Material: uranyl formate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 330 / Average exposure time: 1.0 sec. / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: cryoSPARC / Number images used: 37800 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final 3D classification | Software - Name: cryoSPARC |
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)