[English] 日本語
 Yorodumi
Yorodumi- EMDB-26706: Cryo-EM structure of SHIV-elicited, FP-directed Rhesus Fab RM6561... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
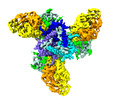
| Title | Cryo-EM structure of SHIV-elicited, FP-directed Rhesus Fab RM6561.DH1021.14 in complex with stabilized HIV-1 Env Ce1176 DS-SOSIP.664 | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | FP / HIV-1 / SOSIP / Vaccine / IMMUNE SYSTEM / Rhesus / SHIV / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / apoptotic process / host cell plasma membrane / structural molecule activity / virion membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of SHIV-elicited, FP-directed Rhesus Fab RM6561.DH1021.14 in complex with stabilized HIV-1 Env Ce1176 DS-SOSIP.664 Authors: Gorman J / Kwong PD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26706.map.gz emd_26706.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26706-v30.xml emd-26706-v30.xml emd-26706.xml emd-26706.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26706.png emd_26706.png | 104.7 KB | ||
| Masks |  emd_26706_msk_1.map emd_26706_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26706.cif.gz emd-26706.cif.gz | 6.8 KB | ||
| Others |  emd_26706_additional_1.map.gz emd_26706_additional_1.map.gz emd_26706_additional_2.map.gz emd_26706_additional_2.map.gz emd_26706_half_map_1.map.gz emd_26706_half_map_1.map.gz emd_26706_half_map_2.map.gz emd_26706_half_map_2.map.gz | 10 MB 31.9 MB 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26706 http://ftp.pdbj.org/pub/emdb/structures/EMD-26706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26706 | HTTPS FTP |
-Validation report
| Summary document |  emd_26706_validation.pdf.gz emd_26706_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26706_full_validation.pdf.gz emd_26706_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_26706_validation.xml.gz emd_26706_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_26706_validation.cif.gz emd_26706_validation.cif.gz | 16.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26706 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26706 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26706 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26706 | HTTPS FTP |
-Related structure data
| Related structure data |  7ur6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26706.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26706.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||
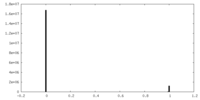
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26706_msk_1.map emd_26706_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
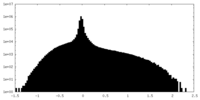
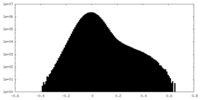
| Density Histograms |
-Additional map: Density modified with resolve
| File | emd_26706_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified with resolve | ||||||||||||
| Projections & Slices |
| ||||||||||||
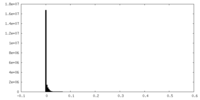
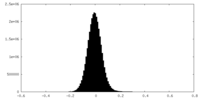
| Density Histograms |
-Additional map: unsharpened map
| File | emd_26706_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
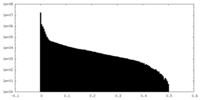
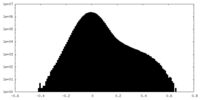
| Density Histograms |
-Half map: half map A
| File | emd_26706_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
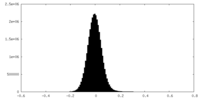
| Density Histograms |
-Half map: half map B
| File | emd_26706_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Fab RM6561.DH1021.14 with stabilized HIV-1 Env Ce1176
| Entire | Name: Fab RM6561.DH1021.14 with stabilized HIV-1 Env Ce1176 |
|---|---|
| Components |
|
-Supramolecule #1: Fab RM6561.DH1021.14 with stabilized HIV-1 Env Ce1176
| Supramolecule | Name: Fab RM6561.DH1021.14 with stabilized HIV-1 Env Ce1176 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: gp120
| Macromolecule | Name: gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.648887 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPAENLWVTV YYGVPVWKEA KTTLFCASDA KAYEKEVHNV WATHACVPTD PNPQEMVLEN VTENFNMWKN DMVDQMHEDV ISLWDQSLK PCVKLTPLCV TLNCTNTTVS NGSSNSNANF EEMKNCSFNA TTEIKDKKKN EYALFYKLDI VPLNNSSGKY R LINCNTSA ...String: GPAENLWVTV YYGVPVWKEA KTTLFCASDA KAYEKEVHNV WATHACVPTD PNPQEMVLEN VTENFNMWKN DMVDQMHEDV ISLWDQSLK PCVKLTPLCV TLNCTNTTVS NGSSNSNANF EEMKNCSFNA TTEIKDKKKN EYALFYKLDI VPLNNSSGKY R LINCNTSA CTQICPKVTF EPIPIHYCAP AGYAILKCNN KTFNGTGPCN NVSTVQCTHG IKPVVSTQLL LNGSLAEKEI II RSENLTN NAKTIIVHLN ESVGIVCTRP SNMTRKSIRI GPGQTFYALG DIIGDIRQPH CNISKQNWNR TLQQVGRKLA EHF PNRNIT FNHSSGGDLE ITTHSFNCRG EFFYCNTSGL FNGTYHPNGT YNETAVNSSD TITLQCRIKQ IINMWQEVGR CMYA PPIAG NITCNSNITG LLLTRDGGIN QTGEEIFRPG GGDMRDNWRS ELYKYKVVEI KPLGIAPTKC KRRVVERRRR RR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: gp41
| Macromolecule | Name: gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.24051 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASNTLTVQA RQLLSGIVQQ QSNLLRAPEA QQHMLQLGVW GFKQLQARVL AIERYLEVQQ LLGIWGCSG KLICCTNVPW NSTWSNRTQE DIWNNMTWME WEREIGNYTH TIYSLLEESQ FQQEINEKDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: Heavy Chain
| Macromolecule | Name: Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.505475 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVTLKESGPA LVKPTQTLTL TCTFSGFSMS NFGSGIYWIR QPPGKALEWL AGIYWTDSKY YNLSLKTRLT ISKDTSKSQV ILIMTNMDP VDTATYYCAD IRGRYYYSNG FLFDVVGVES WGQGVVVTVS S |
-Macromolecule #4: Light Chain
| Macromolecule | Name: Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.679873 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSA SEAARKTVTI SCSGSRSNIG SNSVSWYKQV PGTAPKLLIK YNDQRPSGVS DRFSGSKSGT SASLAISGLQ TEDEADYYC ATWDDSLNGY IFGVGTRLIV L |
-Macromolecule #10: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 10 / Number of copies: 42 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 1.0 X / Component - Formula: PBS / Component - Name: PBS |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 51.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)