+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ClpAP complex bound to ClpS N-terminal extension, class I | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ protease / Adaptor / protein complex / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / protein unfolding / protein catabolic process / peptidase activity / cellular response to heat / ATPase binding / serine-type endopeptidase activity ...endopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / protein unfolding / protein catabolic process / peptidase activity / cellular response to heat / ATPase binding / serine-type endopeptidase activity / ATP hydrolysis activity / proteolysis / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Kim S / Fei X / Sauer RT / Baker TA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: AAA+ protease-adaptor structures reveal altered conformations and ring specialization. Authors: Sora Kim / Xue Fei / Robert T Sauer / Tania A Baker /  Abstract: ClpAP, a two-ring AAA+ protease, degrades N-end-rule proteins bound by the ClpS adaptor. Here we present high-resolution cryo-EM structures of Escherichia coli ClpAPS complexes, showing how ClpA pore ...ClpAP, a two-ring AAA+ protease, degrades N-end-rule proteins bound by the ClpS adaptor. Here we present high-resolution cryo-EM structures of Escherichia coli ClpAPS complexes, showing how ClpA pore loops interact with the ClpS N-terminal extension (NTE), which is normally intrinsically disordered. In two classes, the NTE is bound by a spiral of pore-1 and pore-2 loops in a manner similar to substrate-polypeptide binding by many AAA+ unfoldases. Kinetic studies reveal that pore-2 loops of the ClpA D1 ring catalyze the protein remodeling required for substrate delivery by ClpS. In a third class, D2 pore-1 loops are rotated, tucked away from the channel and do not bind the NTE, demonstrating asymmetry in engagement by the D1 and D2 rings. These studies show additional structures and functions for key AAA+ elements. Pore-loop tucking may be used broadly by AAA+ unfoldases, for example, during enzyme pausing/unloading. #1:  Journal: To Be Published Journal: To Be PublishedTitle: ClpAP AAA+ Protease Adaptor Structures Reveal Pore-Loop Specialization Authors: Kim S / Fei X / Sauer RT / Baker TA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26556.map.gz emd_26556.map.gz | 22.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26556-v30.xml emd-26556-v30.xml emd-26556.xml emd-26556.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26556.png emd_26556.png | 79.6 KB | ||
| Filedesc metadata |  emd-26556.cif.gz emd-26556.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26556 http://ftp.pdbj.org/pub/emdb/structures/EMD-26556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26556 | HTTPS FTP |
-Validation report
| Summary document |  emd_26556_validation.pdf.gz emd_26556_validation.pdf.gz | 528 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26556_full_validation.pdf.gz emd_26556_full_validation.pdf.gz | 527.6 KB | Display | |
| Data in XML |  emd_26556_validation.xml.gz emd_26556_validation.xml.gz | 4.5 KB | Display | |
| Data in CIF |  emd_26556_validation.cif.gz emd_26556_validation.cif.gz | 5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26556 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26556 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26556 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26556 | HTTPS FTP |
-Related structure data
| Related structure data |  7uixMC  7uivC  7uiwC  7uiyC  7uizC  7uj0C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26556.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26556.map.gz / Format: CCP4 / Size: 23.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||
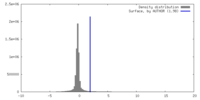
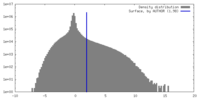
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : AAA+ protease complex ClpAP bound to adaptor protein ClpS and GFP
| Entire | Name: AAA+ protease complex ClpAP bound to adaptor protein ClpS and GFP |
|---|---|
| Components |
|
-Supramolecule #1: AAA+ protease complex ClpAP bound to adaptor protein ClpS and GFP
| Supramolecule | Name: AAA+ protease complex ClpAP bound to adaptor protein ClpS and GFP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent Clp protease ATP-binding subunit ClpA
| Macromolecule | Name: ATP-dependent Clp protease ATP-binding subunit ClpA / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.291758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLNQELELSL NMAFARAREH RHEFMTVEHL LLALLSNPSA REALEACSVD LVALRQELEA FIEQTTPVLP ASEEERDTQP TLSFQRVLQ RAVFHVQSSG RNEVTGANVL VAIFSEQESQ AAYLLRKHEV SRLDVVNFIS HGTRKDEPTQ SSDPGSQPNS E EQAGGEER ...String: MLNQELELSL NMAFARAREH RHEFMTVEHL LLALLSNPSA REALEACSVD LVALRQELEA FIEQTTPVLP ASEEERDTQP TLSFQRVLQ RAVFHVQSSG RNEVTGANVL VAIFSEQESQ AAYLLRKHEV SRLDVVNFIS HGTRKDEPTQ SSDPGSQPNS E EQAGGEER TENFTTNLNQ LARVGGIDPL IGREKELERA IQVLCRRRKN NPLLVGESGV GKTAIAEGLA WRIVQGDVPE VM ADCTIYS LDIGSLLAGT KYRGDFEKRF KALLKQLEQD TNSILFIDEI HTIIGAGAAS GGQVDAANLI KPLLSSGKIR VIG STTYQE FSNIFEKDRA LARRFQKIDI TEPSIEETVQ IINGLKPKYE AHHDVRYTAK AVRAAVELAV KYINDRHLPD KAID VIDEA GARARLMPVS KRKKTVNVAD IESVVARIAR IPEKSVSQSD RDTLKNLGDR LKMLVFGQDK AIEALTEAIK MARAG LGHE HKPVGSFLFA GPTGVGKTEV TVQLSKALGI ELLRFDMSEY MERHTVSRLI GAPPGYVGFD QGGLLTDAVI KHPHAV LLL DEIEKAHPDV FNILLQVMDN GTLTDNNGRK ADFRNVVLVM TTNAGVRETE RKSIGLIHQD NSTDAMEEIK KIFTPEF RN RLDNIIWFDH LSTDVIHQVV DKFIVELQVQ LDQKGVSLEV SQEARNWLAE KGYDRAMGAR PMARVIQDNL KKPLANEL L FGSLVDGGQV TVALDKEKNE LTYGFQSAQK HKAEAAH UniProtKB: ATP-dependent Clp protease ATP-binding subunit ClpA |
-Macromolecule #2: ATP-dependent Clp protease proteolytic subunit
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.660018 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ALVPMVIEQT SRGERSFDIY SRLLKERVIF LTGQVEDHMA NLIVAQMLFL EAENPEKDIY LYINSPGGVI TAGMSIYDTM QFIKPDVST ICMGQAASMG AFLLTAGAKG KRFCLPNSRV MIHQPLGGYQ GQATDIEIHA REILKVKGRM NELMALHTGQ S LEQIERDT ...String: ALVPMVIEQT SRGERSFDIY SRLLKERVIF LTGQVEDHMA NLIVAQMLFL EAENPEKDIY LYINSPGGVI TAGMSIYDTM QFIKPDVST ICMGQAASMG AFLLTAGAKG KRFCLPNSRV MIHQPLGGYQ GQATDIEIHA REILKVKGRM NELMALHTGQ S LEQIERDT ERDRFLSAPE AVEYGLVDSI LTHRNRSHHH HHH UniProtKB: ATP-dependent Clp protease proteolytic subunit |
-Macromolecule #3: ATP-dependent Clp protease adapter protein ClpS
| Macromolecule | Name: ATP-dependent Clp protease adapter protein ClpS / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.193038 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKTNDWLDF DQLAEEKVRD ALKPPSMYKV ILVNDDYTPM EFVIDVLQKF FSYDVERATQ LMLAVHYQGK AICGVFTAEV AETKVAMVN KYARENEHPL LCTLEKA UniProtKB: ATP-dependent Clp protease adapter protein ClpS |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 11 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 6 / Number of copies: 9 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -2.5 µm / Nominal defocus min: -0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7uix: |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)