[English] 日本語
 Yorodumi
Yorodumi- EMDB-26432: Structure of the SARS-CoV-2 S 6P trimer in complex with the neutr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1791 | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Barnes CO | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2022 Journal: Immunity / Year: 2022Title: Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins. Authors: Zijun Wang / Frauke Muecksch / Alice Cho / Christian Gaebler / Hans-Heinrich Hoffmann / Victor Ramos / Shuai Zong / Melissa Cipolla / Briana Johnson / Fabian Schmidt / Justin DaSilva / Eva ...Authors: Zijun Wang / Frauke Muecksch / Alice Cho / Christian Gaebler / Hans-Heinrich Hoffmann / Victor Ramos / Shuai Zong / Melissa Cipolla / Briana Johnson / Fabian Schmidt / Justin DaSilva / Eva Bednarski / Tarek Ben Tanfous / Raphael Raspe / Kaihui Yao / Yu E Lee / Teresia Chen / Martina Turroja / Katrina G Milard / Juan Dizon / Anna Kaczynska / Anna Gazumyan / Thiago Y Oliveira / Charles M Rice / Marina Caskey / Paul D Bieniasz / Theodora Hatziioannou / Christopher O Barnes / Michel C Nussenzweig /  Abstract: SARS-CoV-2 infection or vaccination produces neutralizing antibody responses that contribute to better clinical outcomes. The receptor-binding domain (RBD) and the N-terminal domain (NTD) of the ...SARS-CoV-2 infection or vaccination produces neutralizing antibody responses that contribute to better clinical outcomes. The receptor-binding domain (RBD) and the N-terminal domain (NTD) of the spike trimer (S) constitute the two major neutralizing targets for antibodies. Here, we use NTD-specific probes to capture anti-NTD memory B cells in a longitudinal cohort of infected individuals, some of whom were vaccinated. We found 6 complementation groups of neutralizing antibodies. 58% targeted epitopes outside the NTD supersite, 58% neutralized either Gamma or Omicron, and 14% were broad neutralizers that also neutralized Omicron. Structural characterization revealed that broadly active antibodies targeted three epitopes outside the NTD supersite including a class that recognized both the NTD and SD2 domain. Rapid recruitment of memory B cells producing these antibodies into the plasma cell compartment upon re-infection likely contributes to the relatively benign course of subsequent infections with SARS-CoV-2 variants, including Omicron. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26432.map.gz emd_26432.map.gz | 36.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26432-v30.xml emd-26432-v30.xml emd-26432.xml emd-26432.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26432_fsc.xml emd_26432_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26432.png emd_26432.png | 50.3 KB | ||
| Others |  emd_26432_additional_1.map.gz emd_26432_additional_1.map.gz emd_26432_half_map_1.map.gz emd_26432_half_map_1.map.gz emd_26432_half_map_2.map.gz emd_26432_half_map_2.map.gz | 19.4 MB 35.7 MB 35.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26432 http://ftp.pdbj.org/pub/emdb/structures/EMD-26432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26432 | HTTPS FTP |
-Validation report
| Summary document |  emd_26432_validation.pdf.gz emd_26432_validation.pdf.gz | 816.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26432_full_validation.pdf.gz emd_26432_full_validation.pdf.gz | 816.3 KB | Display | |
| Data in XML |  emd_26432_validation.xml.gz emd_26432_validation.xml.gz | 14.8 KB | Display | |
| Data in CIF |  emd_26432_validation.cif.gz emd_26432_validation.cif.gz | 19 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26432 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26432 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26432.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26432.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.704 Å | ||||||||||||||||||||||||||||||||||||
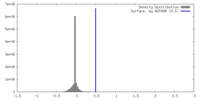
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_26432_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
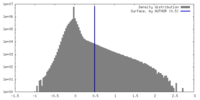
| Density Histograms |
-Half map: #1
| File | emd_26432_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
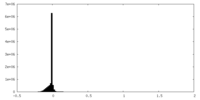
| Density Histograms |
-Half map: #2
| File | emd_26432_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 S 6P + C1791 Fab fragments
| Entire | Name: SARS-CoV-2 S 6P + C1791 Fab fragments |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 S 6P + C1791 Fab fragments
| Supramolecule | Name: SARS-CoV-2 S 6P + C1791 Fab fragments / type: complex / Chimera: Yes / ID: 1 / Parent: 0 Details: Complex between soluble SARS-CoV-2 S 6P bound to C1791 Fab fragments |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: Expi293 Homo sapiens (human) / Recombinant cell: Expi293 |
| Molecular weight | Experimental: 700 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 5206 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)